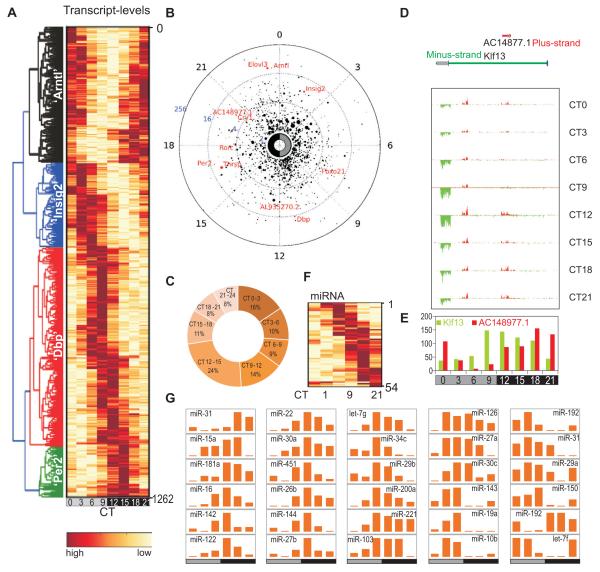
Figure 1. Strand specific and small RNA-Sequencing identified oscillating sense-, antisense-, and mi-RNA transcripts in the mouse liver.
1,262 oscillating transcripts are shown in a (A) heatmap or (B) polar plot. In polar plot, peak-phase is shown along the radial coordinate, the distance to the center of the circle shows the amplitude of transcript oscillation. Average transcript levels over 24 h are proportional to the radius of the marker. Examples of known oscillating transcripts are shown in red. (C) Frequency plot percentage of oscillating transcripts with peak phase of expression binned in 3h over 24h. (D) Klf13/AC148977.1 locus shows oscillations in both sense and antisense transcripts. RNA levels on the plus (red) and minus (green) strands are in 150 bp bins and visualized for each time point. (E) Antisense transcript AC148977.1 (red) and Klf13 (green) are plotted. (F) 54 oscillating miRNAs are shown in a heatmap. (G) Normalized tag counts of example miRNAs show circadian oscillations.