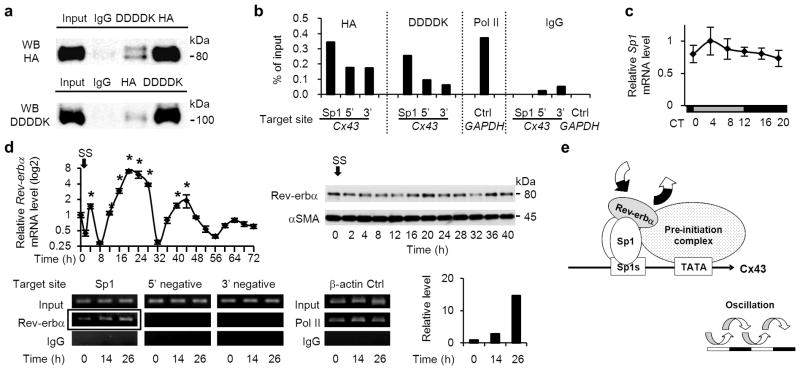
Figure 7. Rhythmic assembly of Rev-erbα and Sp1 at Sp1 sites of the Cx43 promoter.
(a) Co-immunoprecipitation showing a complex formation between HA-tagged Rev-erbα and DDDDK-tagged Sp1 transfected in HEK293T cells, using antibodies for HA and DDDDK. One representative of three experiments with similar results is shown. (b) Chromatin immunoprecipitation (ChIP) assay using antibodies for HA and DDDDK in HEK 293T cells transfected with HA-Rev-erbα and DDDDK-Sp1. Analyses by real-time RT-PCR are shown, targeted against endogenous Sp1 sites of the human Cx43 promoter and its negative control sites, which are approximately 7 kbp up-(5’) and 10 kbp down- (3’) stream from the transcription start site. A ChIP assay using an antibody for RNA polymerase II and primers for human GAPDH promoter was used as a positive control. One representative of two experiments with similar results is shown. (c) Temporal Sp1 mRNA accumulation in the mouse bladder (n=3 for each time point). There were no significant differences among time points by one-way ANOVA. (d) Oscillations of Rev-erbα mRNA (left) and protein expression (right) in serum-shocked rat BSMC (top row). *P < 0.01 vs. the nadir value (time 8) by one-way ANOVA with Dunnett’s post hoc test (n=3–6). SS, serum shock. The ChIP assay, using an antibody for endogenous Rev-erbα, was analysed by RT-PCR targeted against endogenous Sp1 sites of the rat Cx43 promoter and negative control sites, which are approximately 8 kbp up- (5’ negative) and down- (3’ negative) stream (bottom row). β-actin is a positive control. Results of real-time RT-PCR are added; it was targeted against Sp1 sites of the Cx43 promoter, which was immunoprecipitated using an antibody for Rev-erbα (corresponding to the framed bands, bottom). One representative of two experiments with similar results is shown. (e) A mechanistic scheme of Cx43 oscillation, controlled by the Rev-erbα and Sp1 complex binding to Sp1 sites of the Cx43 promoter. Error bars represent s.d. in c and s.e.m. in d. For relative levels, the maximal value was set as 1 in c and the values before serum shock (time 0) was set as 1 in d.