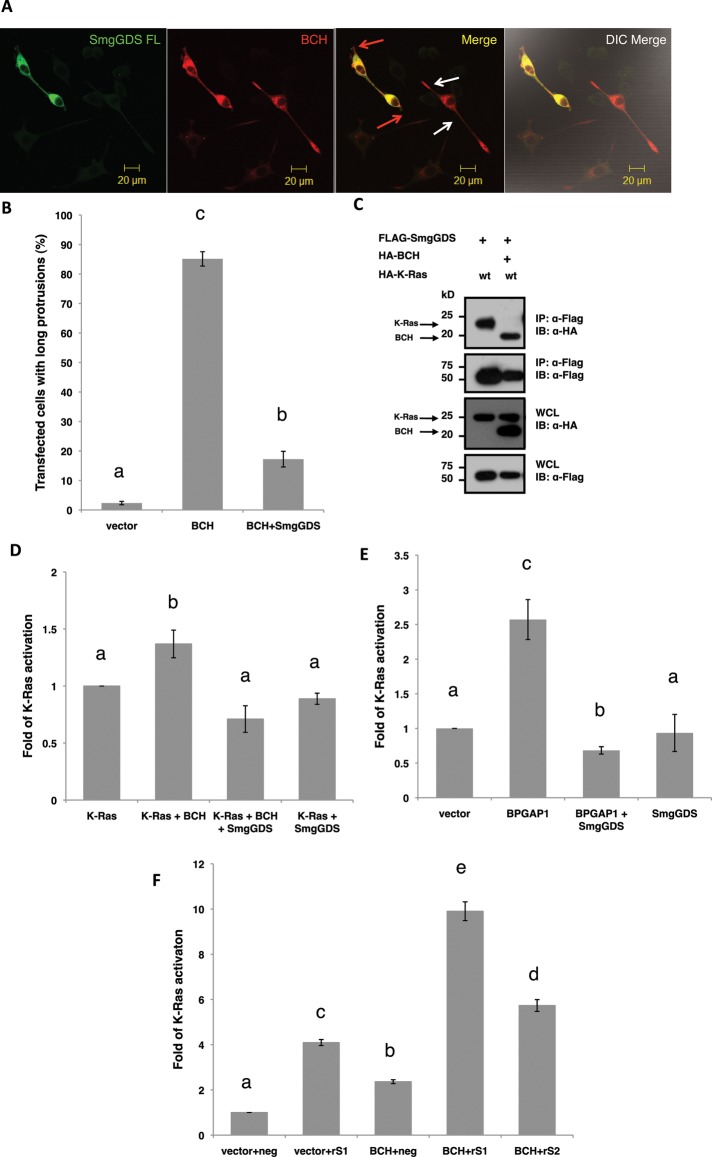
FIGURE 5:
SmgGDS antagonizes BCH domain–induced Ras/ERK activation and neuritogenesis in PC12 cell differentiation. (A) SmgGDS blocks BCH-induced differentiation of PC12 cells. PC12 cells transfected with BCH with or without SmgGDS were made quiescent and stimulated for 48 h with 100 ng/ml EGF before they were fixed for immunofluorescence analysis. Red arrows indicate double-transfected cells, and white arrows indicate single transfection. Scale bars, 20 μm. (B) SmgGDS reduces the percentage of transfected cells with BCH-induced protrusions by ∼70%. Cells were scored for long protrusions (> 2 cell body length) with at least 100 cells counted per sample per experiment. Experiments were performed in triplicate, and the means ± SEM (n = 3), p < 0.01, are plotted. Bars with different letters are highly significant. (C) SmgGDS binds preferentially to BCH over K-Ras, thereby removing BCH from activating K-Ras. HEK293T cells were transfected with SmgGDS and K-Ras either with or without BCH and subjected to IP. (D) SmgGDS blocks BCH activation of K-Ras. Cells transfected with K-Ras, BCH, SmgGDS, or BCH+SmgGDS were subjected to RBD analysis. The intensities of the bound active K-Ras were then quantified using ImageJ, and the means were plotted ± SEM (n = 3). Bars with different letters are significant at p < 0.01. (E) SmgGDS blocks BPGAP1 activation of endogenous K-Ras. Cells transfected with BPGAP1, SmgGDS, or BPGAP1+SmgGDS were subjected to RBD analysis. The intensities of the bound active K-Ras were then quantified using ImageJ, and the means were plotted ± SEM (n = 3). Bars with different letters are significant at p < 0.01. (F) Knockdown of SmgGDS allows a potent activation of K-Ras by BCH. HEK293T cells were transfected with pSilencer negative control (neg), pSilencer rS1 (rS1), singly or together with BCH, or pSilencer rS2 (rS2) together with BCH for 48 h before lysates were subjected to RBD assay. The lysates and bound proteins were then resolved by SDS–PAGE and subsequent Western analysis. Quantified signals were plotted as an average of three independent experiments, as fold of K-Ras activation over the (vector+neg) ± SEM (n = 3). Different letters indicate significance calculated by ANOVA; p < 0.01.