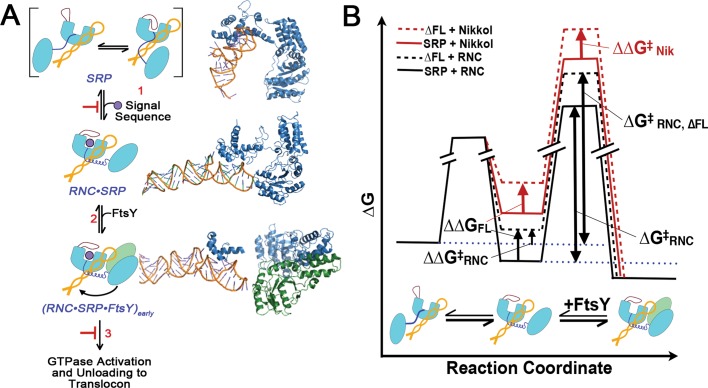
FIGURE 8:
A model for the role of SRP fingerloop (brown) in relaying the information of signal sequence binding to the GTPases and enabling multiple stages of SRP•FtsY interactions during protein targeting is shown in (A). Step 1, the presence of a signal sequence in the M domain is propagated to the NG domain (both in light blue, connected by the flexible linker in dark blue) by the fingerloop, priming the SRP for binding its receptor, FtsY (green), near the SRP RNA's tetraloop end (orange). In step 2, FtsY associates with SRP to form the [RNC·SRP·FtsY]early intermediate. During step 3, [RNC·SRP·FtsY]early rearranges to activate GTP hydrolysis and facilitate the transfer of the RNC to the translocation machinery. Structures or structural models for each complex (PDB IDs [from top to bottom]: 1QZW, 2J28, and 2XKV; Rosendal et al., 2003; Halic et al., 2006; Estrozi et al., 2011) are shown adjacent to the respective SRP/FtsY diagrams. (B) Free-energy profile explaining the different observed effects of fingerloop in the presence of the signal sequence mimic Nikkol (red) or the RNC (black). In both cases, removal of the fingerloop (dashed line) disfavored the conformational change of SRP to an active conformation (ΔΔG) more conducive to complex assembly with FtsY. In the presence of Nikkol, this conformational change is unfavorable, even with wild-type SRP; thus the effect of the fingerloop is fully manifested (ΔΔG = ΔΔG‡Nik). In contrast, when bound to the RNC, the SRP is preorganized into the active conformation; thus, although removal of the fingerloop exerts the same destabilizing effect, the full extent of its defect is masked in the observed complex assembly rates (ΔΔG‡RNC < ΔΔG).