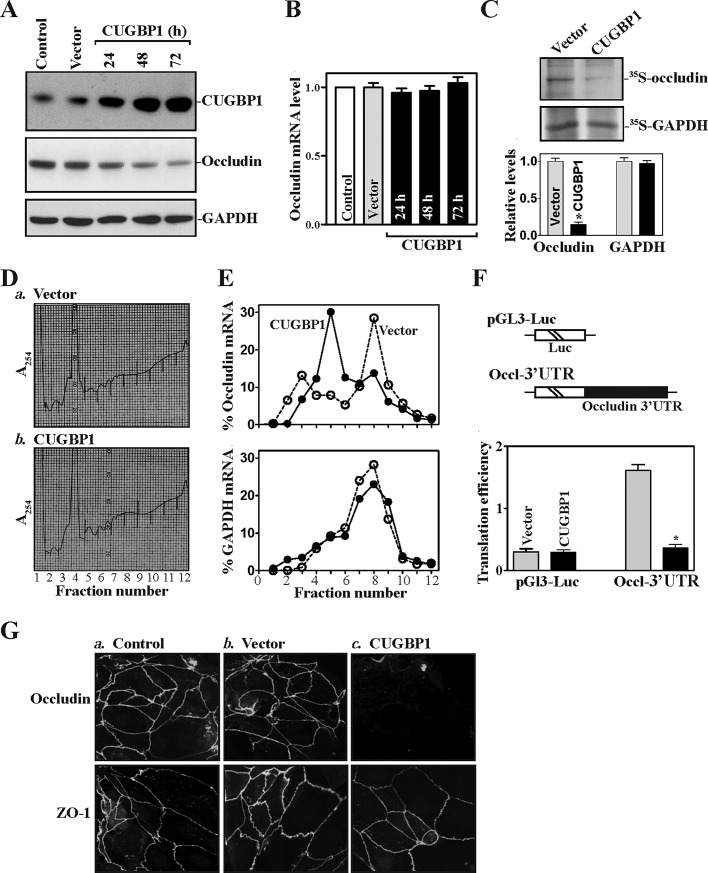
FIGURE 1:
CUGBP1 overexpression inhibits occludin mRNA translation via its 3′-UTR. (A) Representative immunoblots of CUGBP1 and occludin proteins. Cells were transfected with the vector expressing CUGBP1 or control empty vector; protein levels were measured by Western immunoblotting analysis at various times after the transfection by using specific antibody against CUGBP1 or occludin. (B) Levels of occludin mRNA in cells treated as described in panel (A). Total RNA was harvested, and the levels of occludin mRNA were measured by Q-PCR analysis. Data were normalized to GAPDH mRNA levels, and values are shown as the means ± SEM of data from triplicate experiments. (C) Newly translated occludin protein as measured by l-[35S]methionine and l-[35S]cysteine/IP assays in cells transfected with the CUGBP1 expression vector or control vector for 48 h. Top, representative immunoblots of newly synthesized occludin; bottom, quantitative analysis of the immunoblotting signals as measured by densitometry. Values are mean ± SEM of data from three separate experiments. * p < 0.05 compared with cells transfected with control vector. (D) Polysomal profiles in cells described in (C): a) cells transfected with control vector; b) cells transfected with the CUGBP1 expression vector. Nuclei were pelleted, and the resulting supernatants were fractionated through a 10-50% linear sucrose gradient. (E) Distributions of occludin (top) and GAPDH (bottom) mRNAs in each gradient fraction prepared from cells described in panel (D). Total RNA was isolated from the different fractions, and the levels of occludin and GAPDH mRNAs were measured by Q-PCR analysis and plotted as a percentage of the total occludin or GAPDH mRNA levels in that sample. Three independent experiments were performed and showed similar results. (F) Changes in occludin translation efficiency as measured by occludin 3′-UTR-luciferase reporter assays. Top, schematic of plasmids: control (pGL3-Luc); and chimeric firefly luciferase-occludin 3′-UTR (Luc-Occl-3′-UTR). Bottom, levels of occludin translation. The Luc-Occl-3′-UTR or pGL3-Luc (negative control) was cotransfected with a Renilla luciferase reporter. Luciferase values were normalized to luciferase mRNA levels to calculate the translation efficiencies and expressed as means ± SEM of data from three separate experiments. *p < 0.05 compared with cells transfected with control vector. (G) Cellular distribution of occludin and ZO-1 in cells described in (C). After the transfection for 48 h, cells were fixed, permeabilized, incubated with the antibody against occludin or ZO-1, and then with anti-IgG conjugated with FITC. Original magnification, 1000×. Three experiments were performed that showed similar results.