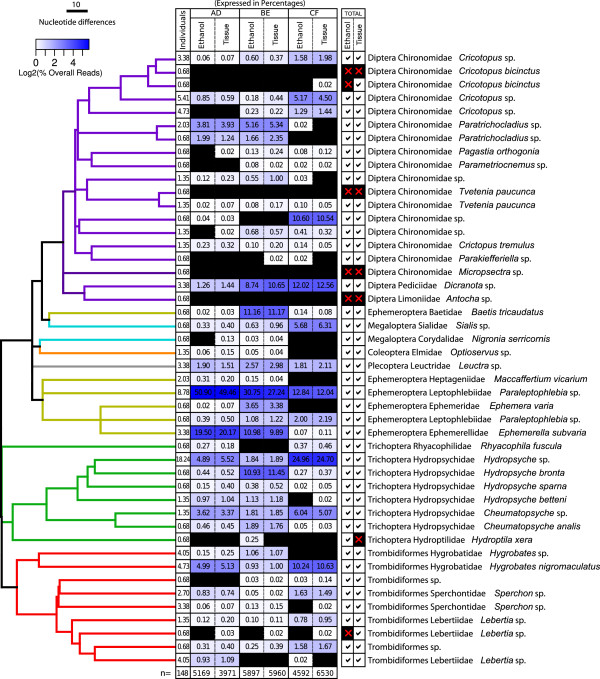
Figure 1.
Taxonomic composition of the benthic sample analysed from tissue-DNA and ethanol-based DNA. The tree is assembled through neighbor-joining analysis of COI DNA barcodes obtained in Sanger sequencing of individual tissue samples and color-coded to visualize different orders. The first column, next to branches, represents percentage of individuals (from a total of 148) for each OTU as identified through Sanger sequencing DNA barcoding. Six subsequent columns represent percentage of sequence reads for taxa in 454 pyrosequencing analysis using the two sources of DNA from mixture (ethanol-based DNA and tissue-DNA) by three primer sets (AD, BE, CF). The final two columns represent total evidence as pass/fail for each taxa using combined data from three primer sets for ethanol-based DNA and tissue-DNA. Cells are color-coded as a heat map based on percentage 454 reads to facilitate visualization of quantitative trends. Black cells represent negative results (no sequence was obtained).