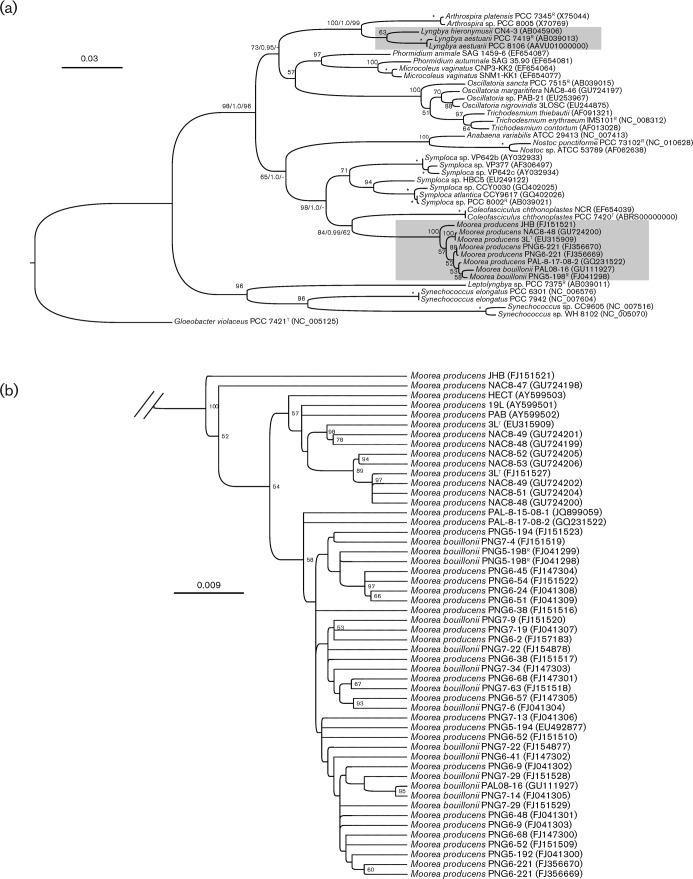
Fig. 1.
(a) Phylogenetic inferences (GARLI) of Lyngbya and Moorea diversification based on SSU (16S) rRNA nucleotide sequences (these two genera are highlighted with shaded boxes). Species and strain numbers of the specimens are given with accession numbers in parentheses. Reference strain (R) and type strain (T) numbers were obtained from Bergey’s Manual (Castenholz, 2001). Support values are indicated as bootstrap and posterior probability for the maximum-likelihood/Bayesian inference/maximum-parsimony methods (* = bootstrap of 100% and a posterior probability of 1.0). Bar, 0.03 estimated nucleotide substitutions per site using the GTR+I+G substitution model. (b) A close-up of the Moorea lineage. Bar, 0.009 expected substitutions per site.