Abstract
The Min system plays an important role in ensuring that cell division occurs at mid-cell in rod-shaped bacteria. In Escherichia coli, pole-to-pole oscillation of the Min proteins specifically inhibits polar septation. This system also prevents polar division in Bacillus subtilis during vegetative growth; however, the Min proteins do not oscillate in this organism. The Min system of B. subtilis plays a distinct role during sporulation, a process of differentiation which begins with an asymmetrical cell division. Here, we show that oscillation of the E. coli Min proteins can be reproduced following their introduction into B. subtilis cells. Further, we present evidence that the oscillatory behaviour of the Min system inhibits sporulation. We propose that an alternative Min system mechanism avoiding oscillation is evolutionarily important because oscillation of the Min system is incompatible with efficient asymmetrical septum formation and sporulation.
Introduction
Rod-shaped bacteria multiply by binary fission, in which the division septum forms with high precision at the cell’s centre. How the division machinery achieves such accuracy is a question of enduring interest. Assembly of FtsZ protomers into a circular structure, called the Z-ring, at the future division site is a prerequisite for cell division (Bi & Lutkenhaus, 1991). It is assumed that initiation of cell division is regulated at the step of FtsZ polymerization and Z-ring placement. Several FtsZ-interacting proteins modulate FtsZ polymerization, acting either to promote or to inhibit this process. MinC prevents FtsZ polymerization and acts as a direct block of polar division (de Boer et al., 1989). In min mutant strains, polar cell division results in the formation of mixtures of ‘mini’ cell forms which lack chromosomes, and extended rods containing multiple nucleoids (Adler et al., 1967; Reeve et al., 1973).
The localization and activity of MinC are dependent on interactions with MinD, an ATPase that associates peripherally with the cytoplasmic membrane (de Boer et al., 1991). MinC and MinD homologues are found in both the Gram-negative Escherichia coli and the Gram-positive Bacillus subtilis. MinD binds reversibly to negatively charged membrane lipids in an ATP-dependent manner (Hu et al., 2002; Hu & Lutkenhaus, 2003; Barák et al., 2008). It is unevenly distributed along the length of the cell, with the highest concentration of MinD and consequently also of MinC found at the cell poles (Marston et al., 1998; Hu & Lutkenhaus, 1999; Raskin & de Boer, 1999a, b). In E. coli, this pattern of localization is determined by MinE. MinE tracks MinD and can be visualized as a ring-like structure at the periphery of the zone occupied by the MinCD complex at the cell pole (Fu et al., 2001; Hale et al., 2001). MinE binding to MinD is accompanied by displacement of MinC and stimulation of the ATPase activity (Hu & Lutkenhaus, 2001), leading to release of MinD from the membrane. Intracellularly, these events lead to net migration of MinD to the opposite cell pole, again followed by MinE, where the molecular events are repeated. This dynamic oscillation process, which takes place with a cycle time of 20–50 s, leads to a MinC concentration minimum at the cell’s centre, where cell division takes place (Hu and Lutkenhaus, 1999; Raskin & de Boer, 1999a, b; Fu et al., 2001; Hale et al., 2001; Juarez & Margolin, 2010; Di Ventura & Sourjik, 2011).
The Min system of B. subtilis features MinC (MinCBs) and MinD (MinDBs), but there is no MinE homologue. Instead, two proteins, MinJ and DivIVA, determine the polar localization of the MinCD complex (Edwards & Errington, 1997; Marston et al., 1998; Bramkamp et al., 2008; Patrick & Kearns, 2008). DivIVA recognizes and binds to negative membrane curvature generated at the newly forming cell poles during cell division, and it recruits the other Min system proteins so as to block the premature formation of a subsequent septum (Lenarcic et al., 2009; Ramamurthi & Losick, 2009; Eswaramoorthy et al., 2011). MinJ, a membrane protein, is recruited by DivIVA to the division site that will become the new cell pole, where it accumulates and serves as a localization signal for MinD (Bramkamp et al., 2008; Patrick & Kearns, 2008).
DivIVA recruits a different set of proteins to the cell poles during sporulation, when it is required for proper segregation of the axial filament, a structure that is composed of elongated sister chromosomes anchored in the vicinity of their ori regions to opposite cell poles (Wu & Errington, 1994, 1998; Webb et al., 1997). In this sporulation-specific chromosomal structure, RacA acts as a bridge between DivIVA at the cell pole and the ori region of the chromosome (Ben-Yehuda et al., 2003; Wu & Errington, 2003). The implied switching of partners by DivIVA may serve to couple relief of inhibition of polar septum formation to faithful chromosome segregation during sporulation. Although deletion of minD has no observable effect on the efficiency of sporulation, the sporulation septum is often misplaced closer to mid-cell in MinD-deficient cells (Barák et al., 1998; Thomaides et al., 2001). At present, the details are not known of how the inhibitory effect of the Min system proteins on polar division is overcome during sporulation.
In B. subtilis, oscillation of the Min proteins has not been observed, indicating a different mechanism of cell division site recognition. Although the Min system in B. subtilis is not as conspicuously dynamic as that in E. coli, there is rapid binding and dissociation of MinDBs molecules at the membrane, and it is postulated that this is accompanied by MinDBs polymerization and depolymerization, respectively (Barák et al., 2008). This characteristic of MinDBs is not so surprising given the high sequence identity between the MinD proteins of B. subtilis and E. coli and the observation of reversible ATP-dependent membrane binding by MinDEc (Drew et al., 2005). The remaining B. subtilis Min system proteins are less dynamic, although rapid movement of MinCBs has been shown following formation of the cell division septum (Gregory et al., 2008).
The different composition and mechanism of action of the Min systems in E. coli and B. subtilis raise interesting evolutionary questions concerning (i) why different mechanisms have evolved to achieve the common goal of disabling polar division, (ii) whether the two mechanisms evolved one from another and, if so, (iii) which of the Min systems appeared first. It is known that MinDEc partially complements MinDBs, and that YFP–MinDEc expressed in B. subtilis localizes on helical trajectories in the same way as GFP–MinDBs (Barák et al., 2008; Pavlendová et al., 2010). This indicates that MinDEc is able to function together with the B. subtilis Min system, and more specifically, to bind to MinCBs (Pavlendová et al., 2010). However, MinE is less promiscuous. It fails to form a ring-like structure or even to localize to the cell membrane of B. subtilis. Instead, the fluorescence signal from MinE–GFP is distributed throughout the cytoplasm, suggesting that the absence of MinE oscillation in B. subtilis is due to its failure to bind to MinDBs.
Here, we show that in the presence of MinE we can reproduce the oscillation of MinDEc in B. subtilis. We also show that cells with oscillating MinD form spores inefficiently. This is not due to defects in signalling, as activation of the response regulator Spo0A occurs normally. Instead, it appears that the cells are affected at the stage of formation of the hallmark of sporulation – an asymmetrical septum. Sporulation would appear therefore to be incompatible with an oscillating Min system, and this may underpin the evolution of different mechanisms in the two bacterial types.
Methods
Bacterial strains, growth conditions and media.
Details of the construction of plasmids and descriptions of B. subtilis and E. coli strains used in this study are presented in Table 1 and Table S1 (available with the online version of this paper), respectively. Sequences of oligonucleotides used in this work are given in Table S2. Strains were grown in Luria broth (LB; Ausubel et al., 1987) or Difco sporulation medium (DSM; Schaeffer et al., 1965) at 37 °C or as stated in the text. DNA manipulations and transformations of E. coli were carried out by standard methods (Sambrook et al., 1989). B. subtilis transformations were performed by the method of Harwood & Cutting (1990). The strains IB1230 and IB1242, with oscillating E. coli Min systems, tend to be unstable. These cells were always freshly prepared by transformation of chromosomal DNA from strain IB1228 into strains IB1111 and IB1112 (Table 1). When required, media were supplemented with the antibiotics ampicillin (100 µg ml−1), tetracycline (10 µg ml−1), kanamycin (10 µg ml−1 or 30 µg ml−1), spectinomycin (100 µg ml−1), chloramphenicol (5 µg ml−1), lincomycin (25 µg ml−1) or erythromycin (1 µg ml−1). Xylose at concentrations of 0.05–0.5 % (w/v) was used for induction of Pxyl; for induction of expression from Phyperspank, 0.1–1 mM IPTG was used.
Table 1. Bacterial strains and their construction.
| Strain | Description | Reference or origin |
| B. subtilis strains | ||
| PY79 | Prototrophic derivative of B. subtilis 168 | Youngman et al. (1984) |
| MO649 | thrC : : cat | Guérout-Fleury et al. (1996) |
| IB220 | spo0A : : kan | Schmeisser et al. (2000) |
| IB1056 | minDBs : : cat | Barák et al. (2008) |
| IB1107 | minDBs : : cat amyE : : Pxyl–minE–gfp spc | Pavlendová et al. (2010) |
| IB1110 | amyE : : Phyperspank–yfp–minDEc spc | Pavlendová et al. (2010) |
| IB1111 | minDBs : : cat amyE : : Phyperspank–yfp–minDEc spc | Pavlendová et al. (2010) |
| IB1112 | minDBs : : cat divIVA : : tet amyE : : Phyperspank–yfp–minDEc spc | Pavlendová et al. (2010) |
| IB1155 | minDBs : : cat amyE : : Phyperspank–yfp–minDEc spc thrC : : Pxyl–minE–gfp erm | IB1111 : : pSGminE |
| IB1244 | trpC2 minJ : : pMUTIN4(bla erm PspaclacZ lacI) minCD : : aph–A3 kan | Bramkamp et al. (2008) |
| IB1228 | thrC : : Pxyl–minE erm | MO649 : : pNP–minE |
| IB1229 | amyE : : Phyperspank–yfp–minDEc spc thrC : : Pxyl–minE erm | IB1110 : : chr DNA IB1228 |
| IB1230 | minDBs : : cat amyE : : Phyperspank–yfp–minDEc spc thrC : : Pxyl–minE erm | IB1111 : : chr DNA IB1228 |
| IB1242 | minDBs : : cat divIVA : : tet amyE : : Phyperspank–yfp–minDEc spc thrC : : Pxyl–minE erm | IB1112 : : chr DNA IB1228 |
| IB1362 | minJ : : kan | PY79 : : pUS19–ΔminJ |
| IB1363 | minDBs : : cat minJ : : kan amyE : : Phyperspank–minDEc spc thrC : : PxylminE erm | IB1230 : : chr DNA IB1362 |
| IB1369 | minCDBs : : kan amyE : : Phyperspank–yfp–minDEc spc | IB1110 : : chr DNA IB1244 |
| IB1370 | minCDBs : : kan amyE : : Phyperspank–yfp–minDEc spc thrC : : PXyl–minE erm | IB1369 : : chr DNA IB1228 |
| IB1371 | minCDBs : : kan | IB333 : : chr DNA IB1244 |
| E. coli strains | ||
| MM294 | F− endA1 hsdR17 (rk−, mk) supE44 thi-1 recA+ | Meselson & Yuan (1968) |
| YLS1 : : pYLS68 | DminCDE Plac : : yfp–minDEc : : minE–cfp | Shih et al. (2002) |
| BTH101 | F− cya-99 araD139 galE15 galK16 rpsL1(Strr) hsdR2 mcrA1 mcrB1 | Karimova et al. (1998) |
Western blotting.
The intracellular levels of GFP, cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) fusion proteins were determined by Western blot analysis with an anti-GFP antibody (Roche Diagnostics) as described previously (Barák et al., 2008). The expression of Spo0A was detected with polyclonal anti-Spo0A antibody. After reaching the stationary phase of growth, cells were collected and processed as described previously (Barák et al., 2008).
Fluorescence microscopy.
Cells were grown to the desired phase and a small amount of culture was transferred to microscope slides covered with a thin layer of 1 % agarose in LB medium. When necessary, cells were concentrated by centrifugation (3 min×2.3 g) and resuspended in a small volume of supernatant prior the examination. To visualize the cells and septal membranes, the cell cultures were stained with FM 4-64 dye (Molecular Probes) at a concentration of 1 µg ml−1. Fluorescence microscopy images were acquired using an Olympus BX61 microscope, equipped with an Olympus DP30BW camera and a spinning disc VivaTome Zeiss microscope. Olympus CellP imaging software and AxioVision 4.8.2.0 software were employed for image acquisition and analysis, and the Huygens Essential software package was used for image deconvolution.
Sporulation efficiency.
The sporulation efficiency was determined essentially as described in Harwood & Cutting (1990). Briefly, cultures were grown in DSM sporulation medium supplemented with 0.5 mM IPTG, 0.5 % xylose and half the dose of appropriate antibiotics at 37 °C for 24 h after inoculation. After heat treatment (85 °C, 15 min), cells were diluted in LB medium and plated onto LB agar plates. Colonies formed from outgrowing spores on these plates represent cells that were able to sporulate and thus survive the heat treatment. These experiments were repeated at least three times. The sporulation efficiency was defined in terms of c.f.u. as follows: (c.f.u. of spores/viable c.f.u.)/(wild-type viable spores/wild-type total viable c.f.u.) and compared with the sporulation efficiency of the wild-type strain, which was taken as 100 %.
Bacterial two-hybrid system.
Fusions of E. coli MinC, MinD and MinE proteins to the T25 and T18 fragments of adenylate cyclase were constructed in the bacterial adenylate cyclase-based two-hybrid (BACTH) system (Karimova et al., 1998). To amplify genes of interest, the primer pairs minCecB2HS and minCecB2HE, minDecB2HS and minDecB2HE or minEecB2HS and minEecB2HE were used with chromosomal DNA from E. coli MM294 strain as template (Meselson & Yuan, 1968). Amplified genes were cloned into the EcoRI and BamHI sites of plasmids pKT25 or pKNT25 and pUT18C or pUT18. Plasmids with T25 and T18 fusions to B. subtilis minC, minD, minJ and divIVA were a kind gift from Dr Richard Daniel, Newcastle University, UK. To test for protein–protein interactions, transformants of E. coli BTH101 (adenylate cyclase-deficient strain) were plated onto LB plates supplemented with 40 µg X-Gal ml−1, 0.1 mM IPTG, 100 µg ampicillin ml−1 and 30 µg kanamycin ml−1, and grown for 24–72 h at 30 °C. To detect interactions, the BACTH system protocol was followed.
Quantitative β-galactosidase assay.
β-Galactosidase activity was measured as described by Miller (1972) with an extra wash step added. To eliminate error due to the effects of different carbon sources in the growth medium, the cells were pelleted and resuspended in an assay buffer prior to further processing.
Results
E. coli MinD oscillation in B. subtilis
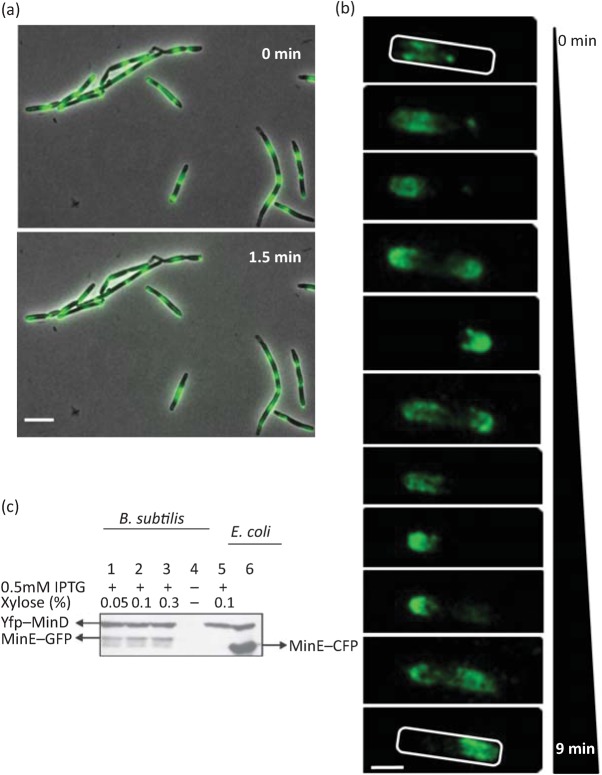
Through a series of genetic manipulations and adjustments to growth conditions, detailed below, we have been able to generate Min system oscillation in B. subtilis. This phenomenon is observed in the majority, if not all, of the cells in the population and occurs with an oscillation cycle time similar to that observed in E. coli (Fig. 1a, Movie S1).
Fig. 1.
E. coli MinD can oscillate in the presence of MinE in B. subtilis. (a) Fluorescence micrographs showing localization of YFP–MinDEc in B. subtilis strain IB1242 (ΔminDBs ΔdivIVA yfp–minDEc minE). In most cells, oscillation of YFP fluorescence could be observed, although in some cells the fluorescence signal appears in the form of dots with reduced mobility. The images were taken with an Olympus BX61 microscope. Two pictures were taken 1.5 min apart. Scale bar, 5 µm. (b) Localization of YFP–MinDEc in a single cell of strain IB1230 (ΔminDBs yfp–minDEc minE). Images were captured using an Olympus BX61 microscope over a period of 9 min and the frames were deconvolved using Huygens Essential software. Scale bar, 1 µm. (c) Relative quantification of YFP–MinD (upper band) and MinE–GFP (lower band, lanes 1–3) in B. subtilis and MinE–CFP (lower band, lane 6) in E. coli by Western blotting. Anti-GFP antibody was used for detection of YFP–MinD, MinE–GFP and MinE–CFP. Lanes 1–3 represent B. subtilis strain IB1155 (ΔminDBs yfp–minDEc minE–gfp) in which expression of yfp–minD is induced with 0.5 mM IPTG and minE–gfp is induced with three different concentrations of xylose, ranging from 0.05 to 0.3 %. Lane 4 represents a negative control, strain IB1056 (ΔminDBs). Lane 5 is strain IB1230 (ΔminDBs yfp–minDEc minE) with expression induced using 0.5 mM IPTG and 0.1 % xylose. Lane 6 represents E. coli strain YLS1 : : pYLS68 grown as described elsewhere (Shih et al., 2002).
MinDEc does not oscillate in the absence of MinE in E. coli (Hu & Lutkenhaus, 2001), nor does it do so when introduced into B. subtilis (Pavlendová et al., 2010). We therefore examined the effect of introducing MinDEc together with MinE into B. subtilis by constructing strains expressing yfp–minDEc and minE in a wild-type (IB1229) and a minDBs deletion (IB1230) background. In many cells, we observed YFP–MinDEc foci close to the cell membrane, especially in strain IB1229. Movement of these ‘dots’ was generally confined to a small local region (Movie S2), and occasionally the dots relocalized towards one of the cell poles. In IB1230 cells, YFP–MinDEc movement reminiscent of oscillation in E. coli was visible, especially in shorter cells (Fig. 1b, Movie S3). Since overexpression of Min proteins causes cell elongation (Marston & Errington, 1999; Pavlendová et al., 2010), the longer cells exhibiting the brightly fluorescing dots are likely to have higher YFP–MinDEc concentrations.
Higher concentrations of MinDEc and MinE may interfere with the function of the Min system by biasing the proportions of the complexes formed. In addition, interaction among E. coli and B. subtilis Min system components may cause slower movement of YFP–MinDEc. In E. coli, the period of the Min oscillation cycle is 20–50 s (Raskin & de Boer, 1999a; Touhami et al., 2006). To compare the oscillation times in E. coli and B. subtilis, we timed the YFP–MinDEc oscillation cycle in E. coli strain ΔminCDE Plac : : yfp–minDEc : : minE–cfp (YLS1 : : pYLS68) (Shih et al., 2002). In our hands, oscillation was observed with a period of about 1 min at room temperature. In contrast, the oscillation of YFP–MinDEc in B. subtilis ΔminDBs yfp–minDEc minE (IB1230) cells was slower at 1.5–3.5 min per cycle. Increasing the temperature to 30 °C, a change that in E. coli results in faster oscillation (from a cycle time of 20 s at 22 °C to 8 s at 30 °C; Touhami et al., 2006), did not significantly enhance the oscillation frequency of YFP–MinDEc in B. subtilis. We reasoned that the presence of B. subtilis DivIVA or MinJ might be limiting the mobility of YFP–MinDEc. To test this idea, we produced YFP–MinDEc and MinE in a B. subtilis strain in which either minD and divIVA (ΔminDBs ΔdivIVA yfp–minDEc minE, IB1242) or minD and minJ (ΔminDBs ΔminJ yfp–minDEc minE, IB1363) were deleted. In these cells, the period of the oscillation cycle was essentially unchanged (1.5–3 min), but oscillation was observed in almost all cells (Fig. 1a, Movie S1).
Next we explored the possibility that the lower frequency of YFP–MinDEc oscillation in the B. subtilis system was caused by perturbations in the concentration ratios of the Min proteins. In the B. subtilis strains described here, YFP–MinDEc and MinE were expressed from the Phyperspank and Pxyl promoters, respectively, while in E. coli YLS1 : : pYLS68, both genes were transcribed from the Plac promoter. To compare MinDE expression levels in B. subtilis and in E. coli, we performed Western blot analysis. It is possible to visualize both MinDEc and MinE on one blot using a monoclonal anti-GFP antibody, in a strain where both MinDEc and MinE are in fusion with fluorescent proteins (ΔminDBs yfp–minDEc minE–gfp, IB1155). Under induction conditions similar to those used for the microscopy experiments (0.5 mM IPTG and 0.1 % xylose), it can be seen in Fig. 1(c) that while the concentrations of YFP–MinDEc (upper bands in lanes 1, 2, 3, 5 and 6) are similar in both systems, the concentration of MinE–CFP (lower band, lane 6) in E. coli strain ΔminCDE Plac : : yfp-minDEc : : minE–cfp (YLS1 : : pYLS68) is higher than the concentration of MinE–GFP in B. subtilis strain expressing both YFP–MinDEc and MinE–GFP (IB1155) (lower band, lanes 1, 2 and 3). Although significant differences in the MinE–GFP expression levels under the three induction conditions tested (Fig. 1c, lanes 1, 2 and 3) were not observed, induction with 0.1 % xylose led to the highest YFP–MinDEc oscillation frequency, which approached one oscillation period per minute in many cells of ΔminDBs (IB1230) and ΔminDBs ΔdivIVA (IB1242) B. subtilis strains. These experiments show that in the presence of MinE, YFP–MinDEc oscillates in B. subtilis and that the characteristics of the oscillation process closely reproduce the oscillation behaviour of the Min system observed in E. coli.
Dynamic MinD inhibits sporulation
Over several days on DSM agar plates, colonies formed by strain ΔminDBs yfp–minDEc minE (IB1230) remained brighter coloured than those formed by wild-type B. subtilis cells, which became darker coloured as the cells sporulated. This suggested that IB1230 cells were impaired in sporulation. We therefore measured the sporulation efficiency of B. subtilis cells expressing the E. coli Min proteins. Interestingly, the sporulation efficiency of strain IB1230 is 10-fold lower (9 %) than that of wild-type cells (Table 2), suggesting that pole-to-pole oscillation of MinD inhibits spore formation. The sporulation efficiency of the strain ΔminDBs ΔdivIVA yfp–minDEc minE (IB1242), which gives the highest YFP–MinDEc oscillation frequency, was not tested, as divIVA mutants are already impaired in sporulation (Thomaides et al., 2001).
Table 2. Sporulation efficiency of B. subtilis strains.
| Strain | Sporulation efficiency | Oscillation | minDBs | minCBs | minDEc | minE |
| PY79 | 100 % | − | + | + | − | − |
| IB1056 | 85±1.9 % | − | − | + | − | − |
| IB1371 | 88.8±0.9 % | − | − | − | − | − |
| IB1111 | 85.4±1.9 % | − | − | + | + | − |
| IB1107 | 56.0±12.0 % | − | − | + | − | + |
| IB1229 | 53.4±17.5 % | +/− | + | + | + | + |
| IB1230 | 8.8±2.5 % | + | − | + | + | + |
| IB1370 | 1.7±0.9 % | + | − | − | + | + |
Next, we inspected cells that retained wild-type minDBs (yfp–minDEc minE, IB1229). The sporulation efficiency of these cells was lower (53 %) than that of the wild-type but significantly higher than that of the ΔminDBs yfp–minDEc minE (IB1230) strain (Table 2). It is possible that higher levels of MinD (MinDBs plus MinDEc) lead to less efficient oscillation and thus to higher sporulation efficiency than that of strain IB1230. To test this possibility, we reinvestigated the sporulation efficiency of strain ΔminDBs yfp–minDEc minE (IB1230) under conditions in which the expression of YFP–MinDEc was increased by addition of 1 mM IPTG. Increased expression of MinDEc had no effect on the sporulation efficiency, which remained at 9 %. It seems therefore that it is the presence of MinDBs per se that causes the partial rescue of sporulation in strain yfp–minDEc minE (IB1229). When we examined the localization of YFP–MinDEc in this strain, we found that in most cells the YFP fluorescence appeared in the form of spots close to the membrane. In comparison with strain ΔminDBs yfp–minDEc minE (IB1230), clear YFP–MinDEc oscillation was visible in fewer cells. This observation implies that MinDBs binds to MinDEc and inhibits its MinE-induced oscillation.
We confirmed the implied interaction between the E. coli and B. subtilis MinD proteins using the bacterial two-hybrid system (Fig. 2). Overall, it seems that oscillation of MinDEc correlates with the lower sporulation frequency. Supporting this assertion, in a control ΔminDBs strain expressing MinDEc in the absence of MinE (ΔminDBs yfp–minDEc, IB1111), where MinDEc does not oscillate, sporulation is unimpaired (Table 2). This result excludes the possibility that the mere presence of MinDEc inhibits sporulation. In a control strain deleted for MinDBs (ΔminDBs, IB1056), the sporulation efficiency is only slightly decreased. In a strain expressing MinE alone (ΔminDBs minE–gfp, IB1107), the sporulation efficiency decreased to 56 %.
Fig. 2.
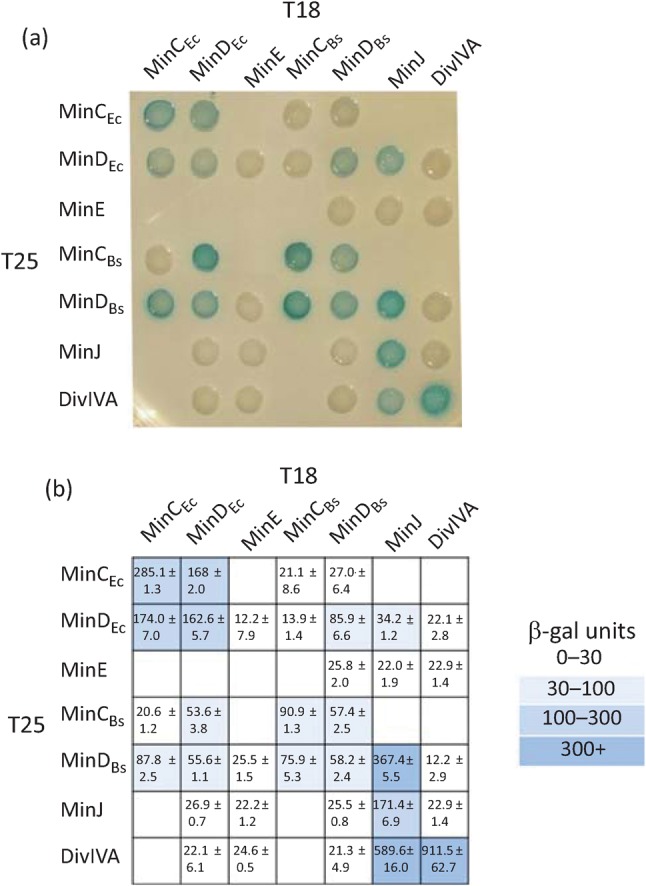
Interactions of Min proteins from E. coli and B. subtilis. E. coli strain BTH101 (Δcya) was co-transformed with plasmids containing the indicated fusions of E. coli and B. subtilis min genes and divIVA to adenylate cyclase fragments T18 and T25. (a) Colonies spotted onto selective X-Gal plates indicate positive (blue) and negative (white) interactions, respectively. (b) The strength of each interaction was quantified by β-galactosidase assay. Numbers indicate Miller units of activity and represent the mean±sd of activity from at least three measurements. Positive interactions are marked by a range of blue colours, as indicated in the key.
The simplest explanation for the decreased sporulation efficiency of strain IB1230 (ΔminDBs yfp–minDEc minE) is that interactions with MinDEc (Fig. 2) induce oscillation of MinCBs, leading to increased MinC concentrations at the cell poles which prevent asymmetrical septation. However, the sporulation efficiency of strain IB1370 (ΔminCBs ΔminDBs yfp–minDEc minE), in which both MinDBs and MinCBs are deleted, is even lower (2 %) than that observed in strain IB1230 (ΔminDBs yfp–minDEc minE). Thus, MinC oscillation does not explain the observed lowering of the sporulation efficiency of strain ΔminDBs yfp–minDEc minE (IB1230), in which MinD oscillation takes place. It is important to note that in MinD-deficient B. subtilis cells, the sporulation septum is often misplaced closer to mid-cell (Barák et al., 1998; Thomaides et al., 2001). In addition, MinCD depletion causes loss of polarity in SpoIIIE-mediated chromosome translocation (Sharp & Pogliano, 2002). However, neither of these two phenotypes is associated with as obvious a reduction in the sporulation efficiency as that observed in strains IB1230 or IB1370 (Table 2). Thus, we assume that the heterologous, oscillating Min system has an additional inhibitory effect on the complex process of sporulation either during asymmetrical septum formation or in the later stages.
Oscillating Min proteins block sporulation by inhibition of polar septum formation
The master regulator of sporulation initiation is Spo0A, a response regulator that is phosphorylated by a multi-component phosphorelay (Hoch, 1993; Perego & Hoch, 2002). Phosphorylated Spo0A binds to specific promoter regions (‘0A boxes’), and activates or represses the expression of scores of genes required for sporulation (reviewed by Piggot & Losick, 2002; Barák et al., 2005). To test whether strains exhibiting oscillation of MinDEc are defective in sporulation initiation, we examined Spo0A expression levels by Western blotting (Fig. 3a). Spo0A is present at similar levels in strain ΔminDBs yfp–minDEc minE (IB1230), which exhibits Min system oscillation, and in the wild-type strain (PY79), which does not. Indeed in all strains examined, Spo0A was detected at normal levels, the exception being the control strain in which spo0A has been deleted (IB220; Schmeisser et al., 2000). Thus the reduced sporulation efficiency associated with the oscillating Min system is not caused by perturbations in the level of Spo0A. Since spo0A expression is positively autoregulated (Molle et al., 2003), normal Spo0A levels indicate that the activity of Spo0A, and the system of proteins that activate it, is unaffected by the oscillating Min system.
Fig. 3.
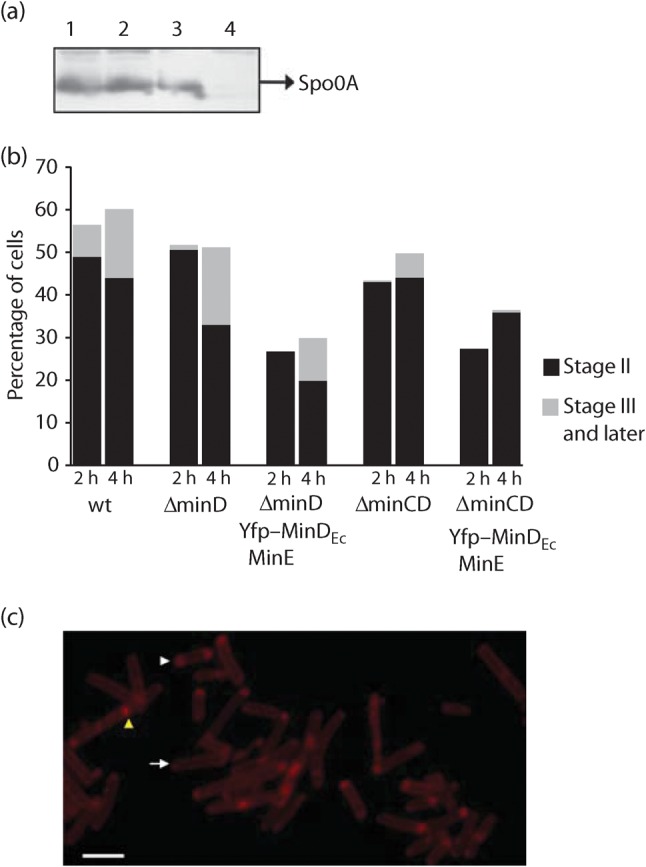
Sporulation block is caused by inefficient asymmetrical septum formation. (a) Western blot with anti-Spo0A antibody, illustrating that the levels of Spo0A in strains where oscillation was observed (IB1242, lane 1; IB1230, lane 2) are similar to levels observed in wild-type B. subtilis strain PY79 (lane 3). This indicates that the block in sporulation is not at the stage of sporulation initiation. No Spo0A was detected in the control strain Δspo0A IB220 (lane 4). (b) To inspect the cells for asymmetrical septum formation, cells were harvested at hour 2 and hour 4 of sporulation, and the membranes were stained using FM4-64 dye. The cells were classified into three groups. First, cells with asymmetrical septa, representing stage II of sporulation (black); cells with a clear minicell phenotype were excluded. Second, cells in later stages of sporulation, stages III and later (grey). The rest of the cells, representing vegetative cells, are not marked. Cells of B. subtilis IB1230 and IB1370 are visibly blocked or delayed in the formation of polar septa. (c) Example of FM4-64-stained cells of IB1370 at hour 2 of sporulation. The arrow indicates a vegetative cell, the white triangle shows a cell in stage II and the yellow triangle a cell in stage III of sporulation. Bar, 3 µm.
A more likely explanation for the lowered sporulation efficiency is a defect in polar cell division. Oscillating MinDEc is expected to bind to MinCBs, thus conferring pole-to-pole oscillation on the cell division inhibitor, which would prevent polar septum formation. This hypothesis was tested by membrane staining. Cells of the wild-type strain (PY79) and strain ΔminDBs yfp–minDEc minE (IB1230) were grown until hours 2 and 4 of sporulation, and the membranes were stained with the dye FM4-64. The pattern of staining defined three discernible cell classes: (i) cells with a polar septum (stage II), (ii) cells in the later stages of sporulation (stage III and later), and (iii) vegetative cells.
For the wild-type strain, after 2 h, 44 % of the cells had not entered into sporulation, 49 % of cells showed a clear polar septum and the remaining cells were in stage III or later (Fig. 3b). Cells of strains ΔminDBs yfp–minDEc minE (IB1230) and ΔminCBs ΔminDBs yfp–minDEc minE (IB1370), which harbour the oscillating E. coli Min system components, were noticeably impaired in the formation of asymmetrical septa. In the second hour of sporulation, forespores in stage III or later were not observed, and an asymmetrical septum was observed in only about 27 % of the cells (IB1230 and IB1370). As mentioned previously, the sporulation efficiency of strain ΔminDBs yfp–minDEc minE (IB1230) is around 9 %. This indicates that even though polar septa are forming in 27 % of these cells at hour 2 of sporulation, only one-third of these give rise to resistant spores. In summary, B. subtilis cells, in which the E. coli Min system proteins oscillate, initiate sporulation normally but are impaired in sporulation septum formation.
Discussion
Regulation of cell division site placement is an intensively studied phenomenon in the model organisms E. coli and B. subtilis. The Min system serves in both classes of organisms as an efficient blockade of unwanted polar septation, but quite different mechanisms of Min system action are postulated. In E. coli, pole-to-pole oscillation of MinCDE creates a concentration gradient of the cell division inhibitor MinC, with the highest concentration at the cell poles, where septation is restricted (Marston et al., 1998; Hu & Lutkenhaus, 1999; Raskin & de Boer, 1999a, b; Hale et al., 2001). In contrast, the MinCDJ–DivIVA complex localizes at the newly formed cell poles and persists at the polar positions in B. subtilis (Edwards & Errington, 1997; Marston et al., 1998; Bramkamp et al., 2008; Patrick & Kearns, 2008, Eswaramoorthy et al., 2011).
The dynamics of MinD localization and reversible membrane binding are integral to the function of both Min systems. The determinant of MinD affinity for the membrane is an amphipathic α-helix at its C terminus (Hu & Lutkenhaus, 2003; Szeto et al., 2003). MinDBs preferentially binds to membranes enriched in negatively charged lipids, such as phosphatidylglycerol, which are helically arranged (Barák et al., 2008). MinDEc also oscillates on a helical trajectory, although it is not known whether helical phosphatidylglycerol domains exist in the cytoplasmic membrane of E. coli (Shih et al., 2003). The phospholipid composition of the membranes of E. coli and B. subtilis is strikingly different. Phosphatidylglycerol represents 40 and 20 % and cardiolipin 24 and 4 % of the membrane phospholipids in B. subtilis and E. coli, respectively (Kusters et al., 1991; López et al., 1998).
These comparisons raise many interesting questions, including whether the E. coli Min system would oscillate following its transplantation into B. subtilis. Elsewhere, oscillation of MinD from Gram-negative Neisseria gonorrhoeae was observed in E. coli (Ramirez-Arcos et al., 2002). Oscillation is an intrinsic property of the Min proteins of E. coli, as shown by the elegant studies on flat membrane systems (Loose et al., 2008). Here we have shown that the E. coli Min system behaves dynamically in Gram-positive B. subtilis. We discovered conditions under which E. coli MinDE oscillation in B. subtilis closely resembles oscillation in E. coli. Oscillation of the Min system proteins is therefore not restricted by the different membrane compositions of E. coli and B. subtilis. This prompts the subsidiary question of why separate mechanisms have evolved to achieve the same goal. One reason could be the incompatibility of Min system oscillation with sporulation. We observed a significant decrease in the sporulation efficiency of B. subtilis cells in which oscillation of E. coli MinD was observed. The defect is not manifested at the stage of sporulation initiation, since expression and activation of the master regulator of sporulation, Spo0A, are unaffected. In contrast, the capacity of the cells to form intact polar septa was impaired, and this was also observed in a strain in which both MinDBs and MinCBs were depleted. Taken together, these results demonstrate that expression of heterologous, oscillating Min proteins restricts polar septum formation by a mechanism that is MinC-independent.
For sporulation to occur there has to be a mechanism for liberating the polar septation sites from the division-inhibitory activity of the Min system. A key factor at this stage is DivIVA, with its alternative functions in vegetative cell division and in sporulation. We speculate that upon binding to RacA, DivIVA loses its capacity to bind to the Min proteins and confine them to the cell poles. This delocalization of the Min proteins would then allow SpoIIE-dependent assembly of FtsZ-rings (Z-rings) at the site of asymmetrical septation. The presence of the oscillating Min system, transplanted from E. coli, has a negative effect on either asymmetrical septum formation or the later stages of the sporulation process, or on both.
Evolution of Min systems and sporulation
The evolutionary implications of these observations are that bacteria which form endospores will have DivIVA/MinJ rather than MinE as the auxiliary component(s) of MinCD. Until recently, sporulation was thought to be restricted to species of Gram-positive bacteria. As shown in Table S3, the genomes of all Gram-positive endospore-forming bacteria encode a DivIVA homologue and most also encode a MinJ homologue. Interestingly, most of the sporulating Clostrideae sp. also possess a MinE homologue (Table S3). However, it is not known whether these MinE proteins are functional, if they are part of Min systems which oscillate, and what, if any, interplay there is with the DivIVA/MinJ system during vegetative growth and sporulation.
The chromosomes of almost all rod-shaped Gram-negative bacteria encode a MinE homologue, and some encode homologues of DivIVA (Rothfield et al., 2005; Table S3). Gram-negative bacteria have hitherto been considered to be non-sporulating, with the possible exception of a sparsely documented example in Thermus. In addition, Myxococcus forms spores by converting the rod-shaped vegetative cell into a spherical spore without prior asymmetrical division (Kaiser, 2003).
From the available data it is hard to deduce which Min system evolved from which, just as we do not know whether the common ancestor of Gram-positive and Gram-negative bacteria possessed these different characteristics. We can speculate that the Min systems either evolved separately or, more likely, evolved together in Gram-positive bacteria for the alternate life cycles of vegetative growth and sporulation, as MinE and DivIVA/MinJ are present in most Clostrideae sp. If this assumption is true, then most probably Gram-negative bacteria evolved from a Gram-positive bacterium. This notion is supported by the recent fascinating description of the cell membrane structures of Acetonoma longum (evolutionarily a close relative of Clostrideae sp.) during sporulation and spore outgrowth (Tocheva et al., 2011). Those authors show that during sporulation the inner membrane of the mother cell is inverted and transformed to become an outer membrane of the germinating cell. Their results point to sporulation as a mechanism by which the bacterial outer membrane could have arisen. If A. longum is the missing link between single- and double-membraned bacteria, it is not surprising that it possesses the two cell-division regulatory systems that characterize Gram-positive and Gram-negative micro-organisms. Further work is needed to address whether and how these two systems function together in the same cell.
Acknowledgements
The authors thank Emília Chovancová for technical assistance, all members of the laboratory for consultations and help, and Lawrence Rothfield (Yu-Ling Shih), University of Connecticut Health Center, Farmington, USA, for E. coli strain YLS1 : : pYLS68 and Richard Daniel, Newcastle University, UK, for bacterial two-hybrid constructs with B. subtilis min genes. We would like also thank Grant Jensen for sharing information about Acetonoma longum ahead of publication. This work was supported by a grant from the Slovak Academy of Sciences (2/0016/10), by a grant from the Slovak Research and Development Agency under contract APVV-00335-10 and by the Welcome Trust project grant 082829.
Abbreviations:
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
Footnotes
Three supplementary tables and three supplementary movies are available with the online version of this paper.
References
- Adler H. I., Fisher W. D., Cohen A., Hardigree A. A. (1967). Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci U S A 57, 321–326. 10.1073/pnas.57.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Siedman J. G., Smith J. A., Struhl K. (1987). Current Protocols in Molecular Biology. New York: Greene Publishing and Wiley. [Google Scholar]
- Barák I., Prepiak P., Schmeisser F. (1998). MinCD proteins control the septation process during sporulation of Bacillus subtilis. J Bacteriol 180, 5327–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barák I., Ricca E., Cutting S. M. (2005). From fundamental studies of sporulation to applied spore research. Mol Microbiol 55, 330–338. 10.1111/j.1365-2958.2004.04445.x [DOI] [PubMed] [Google Scholar]
- Barák I., Muchová K., Wilkinson A. J., O’Toole P. J., Pavlendová N. (2008). Lipid spirals in Bacillus subtilis and their role in cell division. Mol Microbiol 68, 1315–1327. 10.1111/j.1365-2958.2008.06236.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S., Rudner D. Z., Losick R. (2003). RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299, 532–536. 10.1126/science.1079914 [DOI] [PubMed] [Google Scholar]
- Benson A. K., Haldenwang W. G. (1993). Regulation of σB levels and activity in Bacillus subtilis. J Bacteriol 175, 2347–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. F., Lutkenhaus J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164. 10.1038/354161a0 [DOI] [PubMed] [Google Scholar]
- Bramkamp M., Emmins R., Weston L., Donovan C., Daniel R. A., Errington J. (2008). A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol Microbiol 70, 1556–1569. 10.1111/j.1365-2958.2008.06501.x [DOI] [PubMed] [Google Scholar]
- de Boer P. A., Crossley R. E., Rothfield L. I. (1989). A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641–649. 10.1016/0092-8674(89)90586-2 [DOI] [PubMed] [Google Scholar]
- de Boer P. A., Crossley R. E., Hand A. R., Rothfield L. I. (1991). The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J 10, 4371–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ventura B., Sourjik V. (2011). Self-organized partitioning of dynamically localized proteins in bacterial cell division. Mol Syst Biol 7, 457. 10.1038/msb.2010.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D. A., Osborn M. J., Rothfield L. I. (2005). A polymerization-depolymerization model that accurately generates the self-sustained oscillatory system involved in bacterial division site placement. Proc Natl Acad Sci U S A 102, 6114–6118. 10.1073/pnas.0502037102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D. H., Errington J. (1997). The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol 24, 905–915. 10.1046/j.1365-2958.1997.3811764.x [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P., Erb M. L., Gregory J. A., Silverman J., Pogliano K., Pogliano J., Ramamurthi K. S. (2011). Cellular architecture mediates DivIVA ultrastructure and regulates Min activity in Bacillus subtilis. MBio 2, e00257–e11. 10.1128/mBio.00257-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Shih Y. L., Zhang Y., Rothfield L. I. (2001). The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc Natl Acad Sci U S A 98, 980–985. 10.1073/pnas.031549298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory J. A., Becker E. C., Pogliano K. (2008). Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev 22, 3475–3488. 10.1101/gad.1732408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury A. M., Frandsen N., Stragier P. (1996). Plasmids for ectopic integration in Bacillus subtilis. Gene 180, 57–61. 10.1016/S0378-1119(96)00404-0 [DOI] [PubMed] [Google Scholar]
- Hale C. A., Meinhardt H., de Boer P. A. (2001). Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J 20, 1563–1572. 10.1093/emboj/20.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. R., Cutting S. M. (1990). Molecular Biological Methods for Bacillus. Chichester, UK: Wiley. [Google Scholar]
- Hoch J. A. (1993). Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol 47, 441–465. 10.1146/annurev.mi.47.100193.002301 [DOI] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. (1999). Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol 34, 82–90. 10.1046/j.1365-2958.1999.01575.x [DOI] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. (2001). Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell 7, 1337–1343. 10.1016/S1097-2765(01)00273-8 [DOI] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. (2003). A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol 47, 345–355. 10.1046/j.1365-2958.2003.03321.x [DOI] [PubMed] [Google Scholar]
- Hu Z., Gogol E. P., Lutkenhaus J. (2002). Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci U S A 99, 6761–6766. 10.1073/pnas.102059099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju J., Luo T., Haldenwang W. G. (1998). Forespore expression and processing of the SigE transcription factor in wild-type and mutant Bacillus subtilis. J Bacteriol 180, 1673–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez J. R., Margolin W. (2010). Changes in the Min oscillation pattern before and after cell birth. J Bacteriol 192, 4134–4142. 10.1128/JB.00364-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. (2003). Coupling cell movement to multicellular development in myxobacteria. Nat Rev Microbiol 1, 45–54. 10.1038/nrmicro733 [DOI] [PubMed] [Google Scholar]
- Karimova G., Pidoux J., Ullmann A., Ladant D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95, 5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters R., Dowhan W., de Kruijff B. (1991). Negatively charged phospholipids restore prePhoE translocation across phosphatidylglycerol-depleted Escherichia coli inner membranes. J Biol Chem 266, 8659–8662. [PubMed] [Google Scholar]
- Lenarcic R., Halbedel S., Visser L., Shaw M., Wu L. J., Errington J., Marenduzzo D., Hamoen L. W. (2009). Localisation of DivIVA by targeting to negatively curved membranes. EMBO J 28, 2272–2282. 10.1038/emboj.2009.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M., Fischer-Friedrich E., Ries J., Kruse K., Schwille P. (2008). Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792. 10.1126/science.1154413 [DOI] [PubMed] [Google Scholar]
- López C. S., Heras H., Ruzal S. M., Sánchez-Rivas C., Rivas E. A. (1998). Variations of the envelope composition of Bacillus subtilis during growth in hyperosmotic medium. Curr Microbiol 36, 55–61. 10.1007/s002849900279 [DOI] [PubMed] [Google Scholar]
- Marston A. L., Errington J. (1999). Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol 33, 84–96. 10.1046/j.1365-2958.1999.01450.x [DOI] [PubMed] [Google Scholar]
- Marston A. L., Thomaides H. B., Edwards D. H., Sharpe M. E., Errington J. (1998). Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12, 3419–3430. 10.1101/gad.12.21.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M., Yuan R. (1968). DNA restriction enzyme from E. coli. Nature 217, 1110–1114. 10.1038/2171110a0 [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Molle V., Fujita M., Jensen S. T., Eichenberger P., González-Pastor J. E., Liu J. S., Losick R. (2003). The Spo0A regulon of Bacillus subtilis. Mol Microbiol 50, 1683–1701. 10.1046/j.1365-2958.2003.03818.x [DOI] [PubMed] [Google Scholar]
- Patrick J. E., Kearns D. B. (2008). MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol Microbiol 70, 1166–1179. 10.1111/j.1365-2958.2008.06469.x [DOI] [PubMed] [Google Scholar]
- Pavlendová N., Muchová K., Barák I. (2010). Expression of Escherichia coli Min system in Bacillus subtilis and its effect on cell division. FEMS Microbiol Lett 302, 58–68. 10.1111/j.1574-6968.2009.01832.x [DOI] [PubMed] [Google Scholar]
- Perego M., Hoch J. A. (2002). Two component systems, phosphorelays, and regulation of their activities by phophatases. In Bacillus subtilis and its Closest Relatives: from Genes to Cells, pp. 483–517. Edited by Sonenshein A. L., Hoch J. A., Losick R. Washington, DC: American Society for Microbiology Press. [Google Scholar]
- Piggot P. J., Losick R. (2002). Sporulation genes and intercompartmental regulation. In Bacillus subtilis and its Closest Relatives: from Genes to Cells, pp. 473–481. Edited by Sonenshein A. L., Hoch J. A., Losick R. Washington, DC: American Society for Microbiology Press. [Google Scholar]
- Ramamurthi K. S., Losick R. (2009). Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A 106, 13541–13545. 10.1073/pnas.0906851106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Arcos S., Szeto J., Dillon J. A., Margolin W. (2002). Conservation of dynamic localization among MinD and MinE orthologues: oscillation of Neisseria gonorrhoeae proteins in Escherichia coli. Mol Microbiol 46, 493–504. 10.1046/j.1365-2958.2002.03168.x [DOI] [PubMed] [Google Scholar]
- Raskin D. M., de Boer P. A. (1999a). Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci U S A 96, 4971–4976. 10.1073/pnas.96.9.4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D. M., de Boer P. A. (1999b). MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Mendelson N. H., Coyne S. I., Hallock L. L., Cole R. M. (1973). Minicells of Bacillus subtilis. J Bacteriol 114, 860–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L., Taghbalout A., Shih Y. L. (2005). Spatial control of bacterial division-site placement. Nat Rev Microbiol 3, 959–968. 10.1038/nrmicro1290 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. (1965). Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A 54, 704–711. 10.1073/pnas.54.3.704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser F., Brannigan J. A., Lewis R. J., Wilkinson A. J., Youngman P., Barák I. (2000). A new mutation in spo0A with intragenic suppressors in the effector domain. FEMS Microbiol Lett 185, 123–128. 10.1111/j.1574-6968.2000.tb09049.x [DOI] [PubMed] [Google Scholar]
- Sharp M. D., Pogliano K. (2002). MinCD-dependent regulation of the polarity of SpoIIIE assembly and DNA transfer. EMBO J 21, 6267–6274. 10.1093/emboj/cdf597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y. L., Fu X., King G. F., Le T., Rothfield L. (2002). Division site placement in E. coli: mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J 21, 3347–3357. 10.1093/emboj/cdf323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y. L., Le T., Rothfield L. (2003). Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci U S A 100, 7865–7870. 10.1073/pnas.1232225100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto T. H., Rowland S. L., Habrukowich C. L., King G. F. (2003). The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem 278, 40050–40056. 10.1074/jbc.M306876200 [DOI] [PubMed] [Google Scholar]
- Thomaides H. B., Freeman M., El Karoui M., Errington J. (2001). Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev 15, 1662–1673. 10.1101/gad.197501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva E. I., Matson E. G., Morris D. M., Moussavi F., Leadbetter J. R., Jensen G. J. (2011). Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146, 799–812. 10.1016/j.cell.2011.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhami A., Jericho M., Rutenberg A. D. (2006). Temperature dependence of MinD oscillation in Escherichia coli: running hot and fast. J Bacteriol 188, 7661–7667. 10.1128/JB.00911-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C. D., Teleman A., Gordon S., Straight A., Belmont A., Lin D. C., Grossman A. D., Wright A., Losick R. (1997). Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88, 667–674. 10.1016/S0092-8674(00)81909-1 [DOI] [PubMed] [Google Scholar]
- Wu L. J., Errington J. (1994). Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264, 572–575. 10.1126/science.8160014 [DOI] [PubMed] [Google Scholar]
- Wu L. J., Errington J. (1998). Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol 27, 777–786. 10.1046/j.1365-2958.1998.00724.x [DOI] [PubMed] [Google Scholar]
- Wu L. J., Errington J. (2003). RacA and the Soj–Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol 49, 1463–1475. 10.1046/j.1365-2958.2003.03643.x [DOI] [PubMed] [Google Scholar]
- Youngman P. J., Perkins J. B., Losick R. (1984). Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12, 1–9. 10.1016/0147-619X(84)90061-1 [DOI] [PubMed] [Google Scholar]