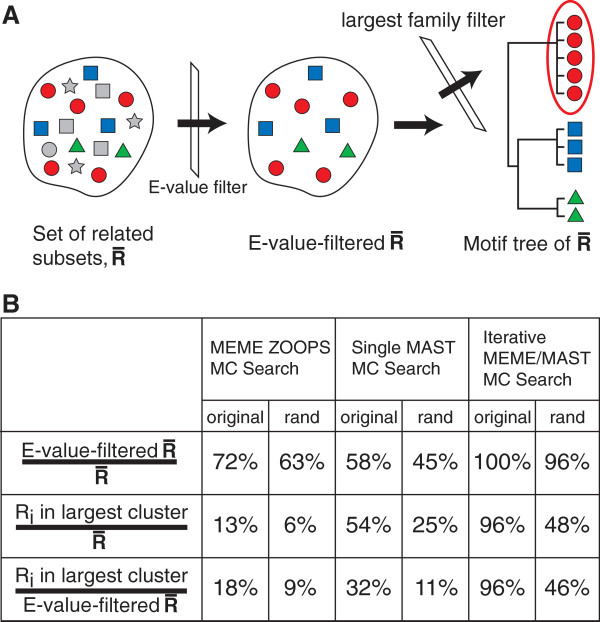
Figure 4.
Motif tree-building protocol and application to ChIP-chip derived LexA binding sites in E Coli. (A) Illustration of MotifCatcher’s motif tree building protocol. In a set of related subsets , all Related Subsets R with associated motifs with an E-value greater than E-value threshold are filtered out (when using the MEME platform or any other motif finder that uses E-value as a measure of statistical significance), and the remaining are organized into a motif tree. The largest motif family describes the motif most likely to be biologically significant. Depending on the input sequence data set, smaller clusters may also represent biologically significant motifs. In this example, Ri-associated motifs are represented as simple polygons (circles, squares, triangles, stars). Gray polygons represent Ri-associated motifs with an E-value above the E-value threshold, and so are eliminated in the initial filtration step. The remaining colored polygons are organized into a motif tree, in which the red circles form the largest cluster (circled in red on tree). In this toy system, there are 17 total , of which 10 pass the E-value filter, of which 5 segregate into the largest cluster. Comparative ratios from the LexA study shown in the table in (B) reveal that regardless of the related subset determination protocol, it was always easier to recover the LexA motif from the original data set versus the set with non-traditional LexA binding sites replaced by random sites. In all trials, the largest cluster motif recapitulated the canonical LexA motif.