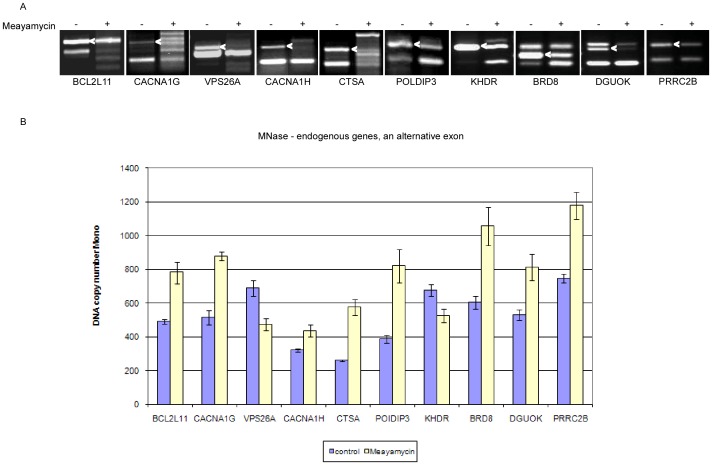
Figure 3. Alternative splicing affects nucleosome occupancy in endogenous genes.
(A) HeLa cells were treated with 10 nM meayamycin and RNA was extracted 24 hr later. Splicing products were separated on a 1.5% agarose gel after RT-PCR reaction using appropriate primers for the flanking the alternative exon (marked with an arrow). Ten endogenous genes were analyzed: BCL2-like 11 (apoptosis facilitator; BCL2L11); calcium channel, voltage-dependent, T type, alpha 1G subunit (CACNA1G); vacuolar protein sorting 26 homolog A (S. pombe) (VPS26A); calcium channel, voltage-dependent, T type, alpha 1H subunit (CACNA1H); cathepsin A (CTSA); polymerase delta interacting protein 3 (POLDIP3); KH domain containing, RNA binding, signal transduction associated 1 (KHDR); bromodomain containing 8 (BRD8); deoxyguanosine kinase, nuclear gene encoding mitochondrial protein (DGUOK); and proline-rich coiled coil 2B (PRRC2B). (B) DNA was extracted from HeLa nuclei 24 hr after meayamycin treatments. An MNase assay was then performed and the mononucleosomal DNA was subjected to absolute QPCR analysis on the alternative exon. Data are presented as DNA copy number.