Figure 3.
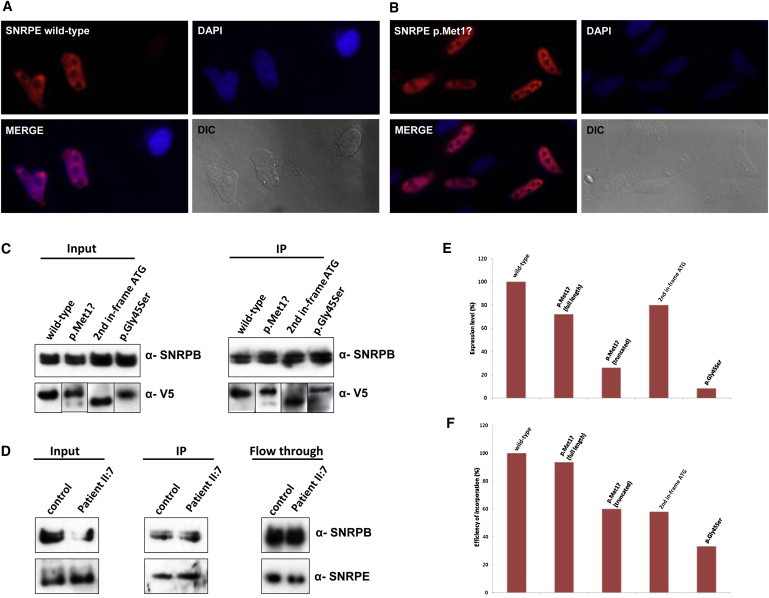
Protein Analyses of SNRPE Wild-Type and Mutant Constructs
(A and B) Subcellular localization of SNRPE wild-type (A) or p.Met1? (B) by immunofluorescence analysis. SNRPE shows a speckled pattern within the nucleus of COS7 cells. In addition, the protein is enriched in discrete spots, which most probably represent Cajal bodies. No significant difference in the localization of the wild-type and mutant SNRPE protein was observed. The nuclei are stained with DAPI; DIC, differential interference microscopy.
(C) Immunoprecipitation of snRNPs with m3G-cap antibody (H20) from transfected HEK293T cells followed by western blot analysis with V5 antibody. Western blotting with SNRPB antibody was used as a control for purification of the snRNPs. We observed that in the case of p.Met1?, both full-length and truncated forms of the protein are translated, albeit at lower levels than the wild-type, and incorporated into the Sm core. We conclude that the initial GTG can be used as a start codon during translation. Integration of the truncated form of the protein (second in-frame ATG) proves that this alteration does not affect the early incorporation of the protein into the Sm core. Interestingly, the substitution p.Gly45Ser is also incorporated into the Sm Core, although with a lower efficiency.
(D) Immunoprecipitation of snRNPs with m3G-cap antibody (H20) from lymphocytes of a control and individual II:7 of the Spanish family (p.Met1?) followed by western blot analysis with SNRPE antibody. Western blotting with SNRPB antibody was used as a control for purification of the snRNPs. Interestingly, in cells of affected individuals, only full-length protein is produced and incorporated into the Sm core with almost equal efficiency to the wild-type. Becauses of identical sizes, one cannot distinguish between wild-type and full-length mutant protein in the cell line of the affected person.
(E) Normalized protein levels of SNRPE wild-type and mutant proteins in HEK293T cells. Quantification has been performed with the results from three separate experiments. In each lane, the protein level has been normalized according to the signal of SNRPB in the same lane and been shown as a percentage of the wild-type protein. Quantifications have been performed by the ImageJ program.
(F) Normalized efficiency of incorporation of wild-type and mutant SNRPE proteins into the Sm core in HEK293T cells. The efficiency of incorporation in each lane has been normalized according to the signal of SNRPB in the same lane and also the protein level in the same experiment. Quantification has been performed with the results from three separate experiments by the ImageJ program.
