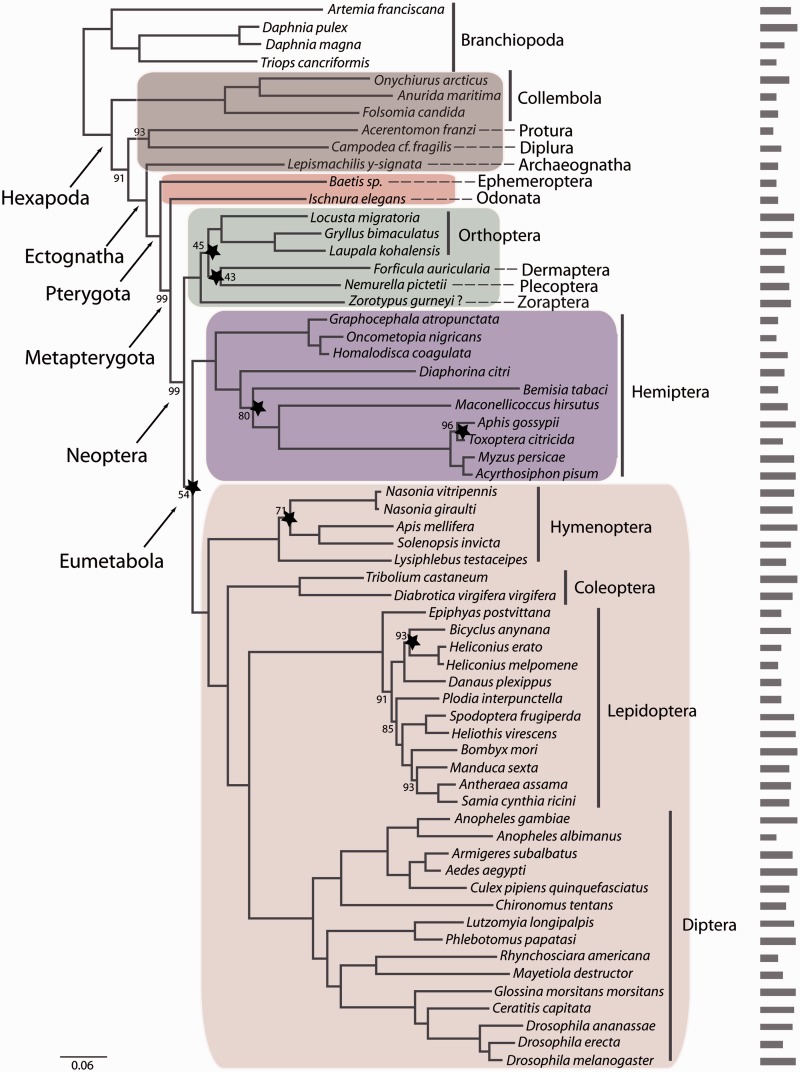
Fig. 2.—
RAxML topology derived from data matrix P_matrix_g (62 species, 285 genes, 79506 amino acid positions), PROTCATWAGF. Support values are derived from 1,000 bootstrap replicates. Bootstrap values are only given for nodes that lack maximum support. Stars indicate nodes for which at least 90% of data (=256 genes) have to be concatenated to recover this specific node based on the RADICAL analyses. Also shown is a barplot indicating the number of putative orthologous genes for each of the taxa in this data set. Color code: primarily wingless hexapods, gray; “palaeopteran” insects, red; polyneopteran insects, green, paraneopteran insects, purple; and holometabolan insects, pink.