Abstract
The relative importance of mutation, selection, and biased gene conversion to patterns of base composition variation in Drosophila melanogaster, and to a lesser extent, D. simulans, has been investigated for many years. However, genomic data from sufficiently large samples to thoroughly characterize patterns of base composition polymorphism within species have been lacking. Here, we report a genome-wide analysis of coding and noncoding polymorphism in a large sample of inbred D. melanogaster strains from Raleigh, North Carolina. Consistent with previous results, we observed that AT mutations fix more frequently than GC mutations in D. melanogaster. Contrary to predictions of previous models of codon usage in D. melanogaster, we found that synonymous sites segregating for derived AT polymorphisms were less skewed toward low frequencies compared with sites segregating a derived GC polymorphism. However, no such pattern was observed for comparable base composition polymorphisms in noncoding DNA. These results suggest that AT-ending codons could currently be favored by natural selection in the D. melanogaster lineage.
Keywords: synonymous codon, genome evolution, mutational bias, natural selection
Introduction
The evolution of codon usage bias has been studied extensively in many organisms, notably prokaryotes (Shah and Gilchrist 2010; Supek et al. 2010) and Drosophila (Akashi 1994, 1995, 1996; Heger and Ponting 2007). Codon bias may result from mutation bias or from natural selection (Stenico et al. 1994; Akashi et al. 1998; Singh et al. 2007). The strength and effect of selection on codon bias are often inferred from genomic patterns of codon usage or from divergence between species. For example, in Drosophila, genes that show more biased codon usage are enriched for GC-ending codons (Akashi 1994; Duret and Mouchiroud 1999) and tend to be expressed at higher levels. It is inferred from these patterns and from the fact that noncoding DNA is considerably more AT-rich than synonymous sites (Shields et al. 1988; Moriyama and Hartl 1993) that GC-ending codons in Drosophila are favored (i.e., preferred) by natural selection, presumably owing to selection on translational efficiency or accuracy (Ikemura 1982; Akashi 1994; Duret 2002). It is also conceivable that the frequency of derived preferred codons, which serve as advantageous mutations in highly expressed genes, would be elevated by natural selection (Akashi and Schaeffer 1997). Moreover, several studies have suggested that effects of synonymous site variation on protein folding could be an agent of natural selection for codon bias (Kimchi-Sarfaty et al. 2007; Stoletzki and Eyre-Walker 2007; Drummond and Wilke 2008). Recent genomic sequencing and annotation of several Drosophila species genomes suggested that genomic patterns of GC biased codon usage are widely shared within the genus. Nevertheless, the recent evolution of codon usage in the Drosophila saltans complex shows that major evolution of codon usage could occur (Rodriguez-Trelles et al. 1999; Singh et al. 2006). In addition to data on genomic patterns of base composition and divergence, functional experiments in Drosophila also show fitness effects of synonymous site variation. For example, transgenic flies carrying a manipulated version of alcohol dehydrogenase with unpreferred codons exhibited altered ethanol tolerance (Carlini 2004; Hense et al. 2010).
Analysis of Drosophila codon usage variation on shorter time scales has revealed a composite view on the codon usage evolution. Patterns of polymorphism and divergence in D. melanogaster suggested that recent selection on the codon usage in this species is either very weak (comparing with selection on D. simulans and their ancestral lineage) or absent (Akashi 1995; Andolfatto 2007; Nielsen et al. 2007; Singh et al. 2007). One explanation is that a recent population size decrease in D. melanogaster has enhanced the role of genetic drift (Akashi 1997; McVean and Vieira 2001) relative to D. simulans. However, the simple model in which D. melanogaster is approaching a new equilibrium under mutation, selection, and drift may be incorrect. For example, several Drosophila lineages, including D. simulans appear to be evolving in codon usage (Akashi 1996; Begun and Whitley 2002) suggesting that the codon usage evolution observed in D. melanogaster is common, and recent results cast doubt on the idea that the long-term effective population size is very different in D. melanogaster versus D. simulans versus the D. melanogaster/D. simulans ancestor (Nolte and Schlotterer 2008). In addition, several studies provided evidence that unpreferred codons which in general are thought to be deleterious have been driven to fixation by natural selection at some sites in the D. melanogaster lineage (Bauer DuMont et al. 2004; Nielsen et al. 2007; Singh et al. 2007; Holloway et al. 2008), suggesting that selection coefficients on synonymous mutations may vary across the genome and be context dependent (Nielsen et al. 2007; Singh et al. 2007). One approach for investigating the mutation–selection–drift model is the analysis of the frequency spectrum of polymorphism and the comparison of polymorphism to divergence. Although there have been some studies on population genetics of codon usage variation in Drosophila (Akashi 1994, 1995; Begun 2001; Kern and Begun 2005; Galtier et al. 2006; Begun et al. 2007; Haddrill and Charlesworth 2008; Zeng and Charlesworth 2010), the data sets have been small in terms of numbers of sites and/or alleles, which is a serious drawback for investigating weak selection. To investigate the population genetics of codon usage variation, we took advantage of a large collection of genomic sequences from a Raleigh, North Carolina, population of D. melanogaster.
Materials and Methods
Data Set
The population genomic sequences of D. melanogaster were produced by DPGP (dpgp r1.0, http://www.dpgp.org/1K_50genomes.html#Reference_Release_1.0, last accessed December 4, 2012). The description of the sequencing, alignment, and assignment of estimated quality scores is described in Langley et al. (2012). Each base was filtered with a minimum quality score of Q30 necessary to be included in these analyses. Langley et al. (2012) established a conservative gene set that comprised genes for which all sequenced D. melanogaster alleles agreed to the reference sequence start codon, splice junctions, and termination codon. Genes having alignments less than 33 codons were removed from the analysis. The 9,328 genes that satisfied the above criteria were included in the following analyses. The noncoding DNA data set was derived from introns and intergenic regions; nucleotides that overlapped a coding region in any mRNA isoforms were removed from our noncoding data set. Because of the limited number of sequenced Malawi strains, most of the analyses were based on the Raleigh samples.
Substitution Rate Estimation
Substitutions on the D. melanogaster and D. simulans lineages were polarized based on parsimony; D. yakuba (or D. erecta if D. yakuba sequence is not available) was used as an outgroup to infer the D. melanogaster/D. simulans ancestral state. Sites with a gap or undetermined base in either species were removed from the analysis. To estimate the substitution rate with respect to base composition for a region, the number of substitutions was counted and then divided by the number of appropriate sites in the ancestral sequence to obtain the substitution rate. In other words, the denominator used in estimating the AT substitution rate is the number of inferred ancestral GC sites. The log2 ratio of substitution rates of AT-to-GC (GC mutations) to that of GC-to-AT (AT mutations) was calculated as the index of substitution rate bias. Because the index comprised the ratio of substitution rates but not numbers, the index should not be biased if AT/GC nucleotides are not of equal amount. A positive index denotes GC over AT preference, whereas a negative value indicates the opposite.
Recombination Rate Estimation
The local recombination rate based on the physical location was estimated by fitting a curve in the Marey maps based on the empirical data from D. melanogaster (Fiston-Lavier et al. 2010). Genes were then grouped into recombination categories according to Singh et al. (2005) as follows: N: none, recombination less than 0.27 cM/Mb, and 939 genes are in this class; L: low, between 0.27 and 2.93 cM/Mb, and 3,320 genes are in this class; M: medium, recombination between 2.93 and 3.90 cM/Mb, and 3,585 genes are in this class; and H: high, recombination greater than 3.9 cM/Mb and 986 genes are in this class. Because the number of polymorphisms for “N” and “L” was too small to test properties of the frequency spectrum (see later), sites in the “N” and “L” categories were combined and considered as low-recombination sites for these analyses. Similarly, data corresponding to “M” and “H” were also combined and considered as high-recombination rate genes for comparison.
Expression Level
We used the Affymetrix expression microarray data in FlyAtlas (http://flyatlas.org/, last accessed December 4, 2012, Chintapalli et al. 2007) as estimates of expression level. The data from the four biological replicates of whole fly RNA preparations were averaged. The highly expressed genes were defined as those yield signal greater than 200, and the lowly expressed genes were those with signal less than 50. The definition was followed by the online description.
Polymorphism Analysis
For the Raleigh and Malawi samples, sites with a sample size less than 30 and 5, respectively, were filtered out. Drosophila melanogaster polymorphisms were polarized using the outgroup sequences, D. simulans and D. yakuba. All sites that were variable in D. melanogaster and in the outgroup were excluded from this analysis. For coding region comparisons, only synonymous sites are considered, and only codons with a single synonymous change in the population were retained. Contingency table tests (analogous to McDonald–Kreitman tests) were performed by comparing ratios of GC to AT polymorphic and fixed variants. χ2 was used as a test for statistical significance. Percentage deviation for these tables was estimated as a measure of the degree to which an observed χ2 cell departed from the expected value under the null hypothesis and was calculated as the difference of observed and expected value divided by expected value and then multiplied by 100%. Tajima’s D (Tajima 1989) and Fu and Li’s D* (Fu and Li 1993) were used as estimators of skewness of the site frequency spectra. The significance level of D and D* were determined by using ms (Hudson 1990, 2002) to simulate a neutral equilibrium population with theta equal to the observed values and recombination rate of zero. We used only sites with coverage of 35 alleles for the frequency spectra analyses, though the results are similar for sites with lower coverage (supplementary table S3, Supplementary Material online) and more stringent quality score (supplementary table S4, Supplementary Material online). The comparison of the full frequency spectrum across mutation types was performed by the Mann–Whitney U (MWU) test. The MWU test was performed by comparing the number of SNPs of each derived allele frequency between AT and GC mutations.
Results
Divergence
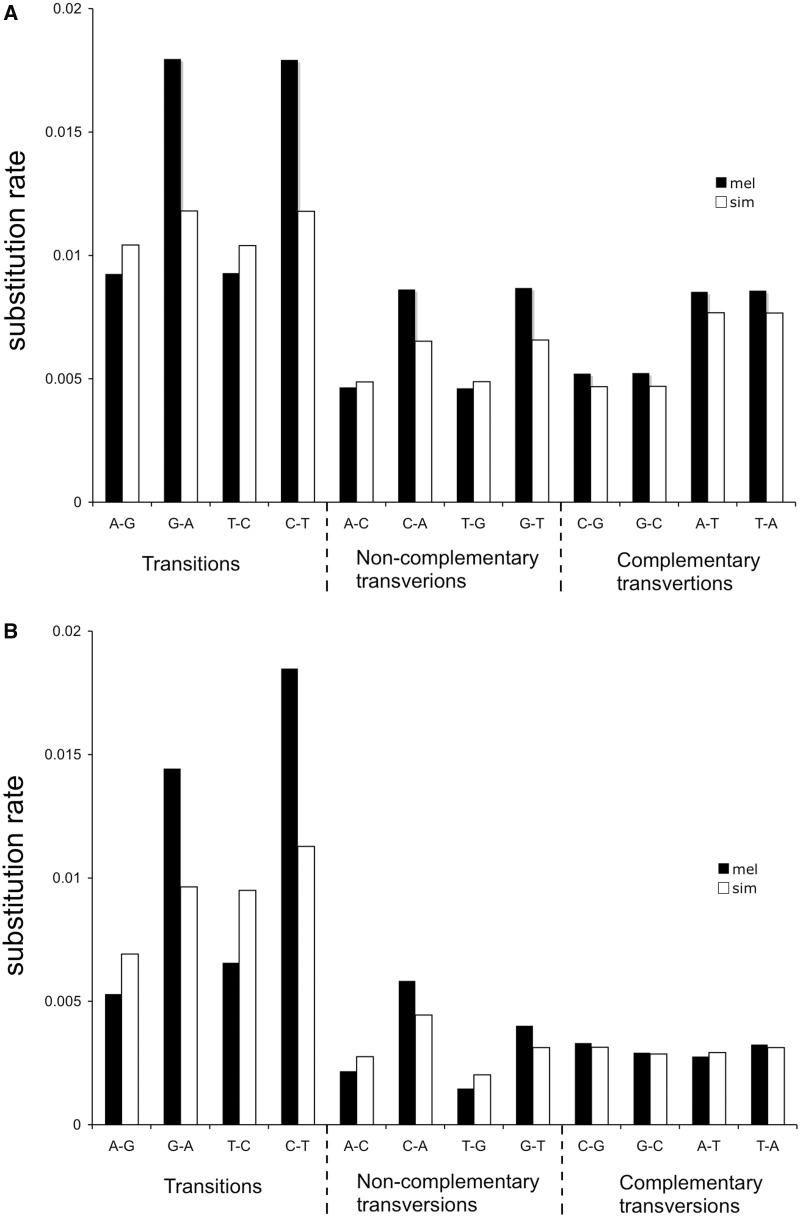
The substitution rates for base composition variants in D. melanogaster and D. simulans were highly heterogeneous, especially for the transitions and noncomplementary transversions. The whole genome GC-to-AT substitution rate based on whole genome (herein referred to as the AT substitution rate) was higher than the GC substitution rate in both lineages, but the bias was more extreme in D. melanogaster. For example, the substitution rate for G → A was almost twice (0.0180/0.0093) the rate for A → G in D. melanogaster (fig. 1A, G → A: 0.0180; A → G: 0.0093, G-test P < 0.001) but was only 13% higher in D. simulans (fig. 1A, G → A: 0.0118; A → G: 0.0104, G-test P < 0.01). Coding regions also exhibited AT substitution bias in both D. melanogaster and D. simulans and again was more extreme in D. melanogaster. As shown in figure 1B, the substitution rate for G → A was 2.72 times the rate for A → G in D. melanogaster coding region and increased dramatically compared with the substitution rate estimated from whole genome (1.94X). However, the AT-increasing substitution rates are similar for coding regions (1.36X) and the whole genome (1.13X) in D. simulans. Intriguingly, the AT fixation biases were even more extreme for the X chromosome (supplementary fig. S1, Supplementary Material online). These results regarding AT substitution bias are concordant with previous studies (Akashi 1996; Takano-Shimizu 2001; Begun and Whitley 2002; Kern and Begun 2005; Akashi et al. 2007). Our genomic estimate of GC-to-AT substitution bias (1.914X), which primarily comprised noncoding DNA, is comparable to the mutation bias (2.0X) inferred from sequencing of mutation accumulation lines (Keightley et al. 2009). The much larger ratio observed for coding regions (2.385X) suggests that mutation bias itself cannot be the only explanation.
Fig. 1.—
Substitution rate of different mutation classes. (A) The average substitution rate for all autosomal genome in Drosophila melanogaster and D. simulans. (B) The average substitution rate for autosomal-linked coding genes in D. melanogaster and D. simulans.
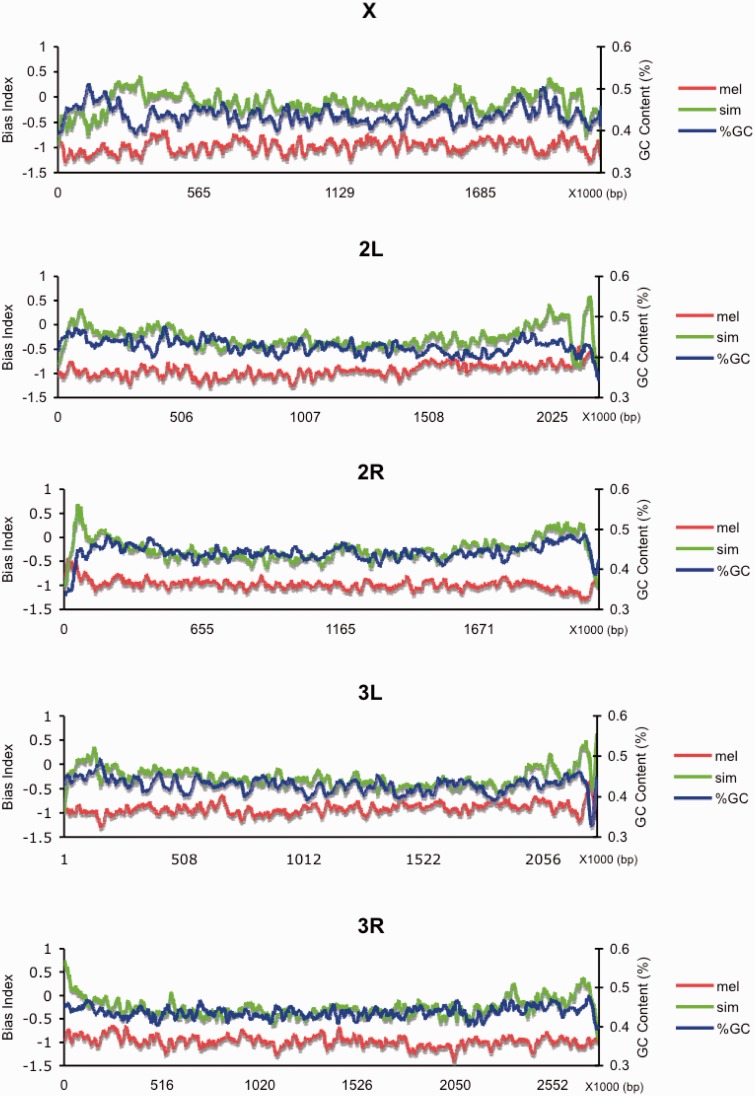
Given that the D. melanogaster lineage exhibits a strong AT fixation bias, it is natural to ask whether the degree of bias is heterogeneous across the genome. The lineage-specific substitution bias (see Materials and Methods) was calculated for 10 kb sliding windows in both D. melanogaster and D. simulans (fig. 2). In D. melanogaster, AT was fixed much more frequently than GC in almost all the windows surveyed. Though D. simulans showed less AT substitution bias than D. melanogaster; there was a broad and significant genomic trend in D. simulans toward slightly negative substitution bias indicative of increased AT fixation, consistent with previous analyses (Akashi 1997; Begun et al. 2007). The level of AT substitution bias was negatively correlated with ancestral GC content in D. melanogaster but positively correlated in D. simulans (Spearman’s ρ = −0.1258 for the melanogaster lineage, P < 0.001; ρ = 0.1066 on the simulans lineage, P < 0.001, fig. 2). In other words, the D. melanogaster genome has evolved toward higher AT content and has evolved more in ancestrally GC-rich regions. Although there is a slight tendency toward evolving higher AT content in D. simulans, regions that possess higher GC ancestrally showed less accumulation of AT substitutions.
Fig. 2.—
Genomic plot of substitution bias. Sliding-window analysis of the substitution bias calculated per 10 kb regions for Drosophila melanogaster (red) and D. simulans (green) genomic sequences, using D. yakuba as outgroup. GC content is derived from the interpreted common ancestor of D. melanogaster and D. simulans common ancestor (blue).
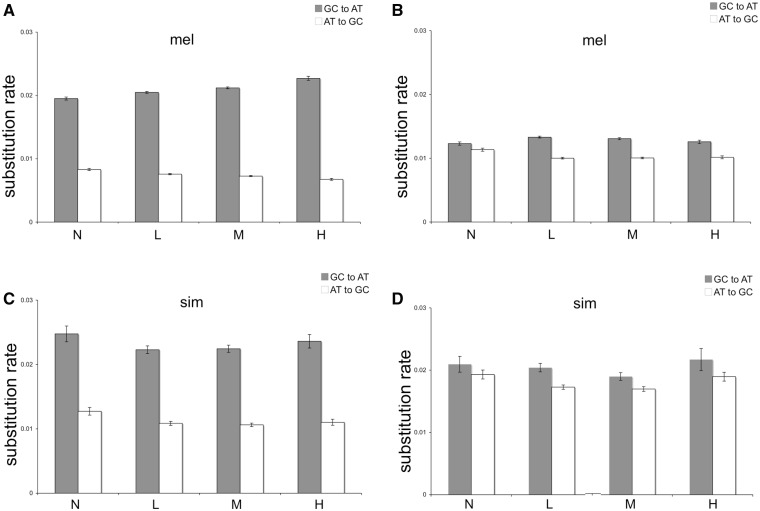
Under the mutation–selection–drift model of codon bias evolution (Sharp and Li 1986; Bulmer 1991), AT-ending codons are weakly deleterious and are more likely to be fixed in regions of lower recombination, whereas GC-ending codons are slightly beneficial and more likely to be fixed in regions of higher recombination owing to Hill–Robertson effects (Hill and Robertson 1966; Comeron et al. 1999). This model predicts a positive correlation between recombination and substitution bias. Similarly, if recombination and GC-biased gene conversion are correlated (Marais et al. 2003), a positive correlation between recombination rate and GC substitution bias would be expected. However, we observed a negative correlation between recombination and substitution bias at D. melanogaster synonymous sites (Spearman’s ρ = −0.1096, P < 0.001; fig. 3) which is in agreement with earlier results (Bauer DuMont et al. 2009). In contrast to the pattern at synonymous sites, there was less AT substitution bias observed in intronic sequences, and the correlation between recombination and substitution bias is relatively subtle (Spearman’s ρ = −0.035, P = 0.0082), which suggests that gene conversion is not the major factor driving the observed substitution bias. In short, the results from synonymous sites and intron sites suggest little role for regional effects of mutation bias or recombination in D. melanogaster contributing to the observed correlations between substitution bias and crossing over (perhaps gene conversion). In D. simulans, we observed no correlation between synonymous site substitution bias and recombination (Spearman’s ρ = −0.0143, P = 0.0836), though we did observe a weak positive correlation between recombination and substitution bias in introns (Spearman’s ρ = 0.038, P < 0.0056). The analyses in D. simulans may be compromised by the fact that recombination rate estimates are from D. melanogaster.
Fig. 3.—
Relationship between recombination rate and the profile of base composition evolution. Mean substitution rate (±Standard Error) of AT and GC nucleotide substitutions of Drosophila melanogaster (A: exon and B: intron) and D. simulans genome (C: exon and D: intron). Within each data set, the two types of substitutions were subdivided into categories according to frequency of crossing over: high (H), >3.9 cM/Mb; medium (M), 2.93–3.9 cM/Mb; low (L), 0.27–2.93 cM/Mb; and none (N), <0.27.
Polymorphism
To examine base composition evolution in D. melanogaster on a shorter time scale, the noncoding and synonymous fixations were compared with polymorphism. If base composition variation is neutral, the ratio of AT-to-GC versus GC-to-AT polymorphisms should be similar to the ratio of AT-to-GC versus GC-to-AT fixations. The data show significant deviations from this expectation (all P value <0.01, in table 1, supplementary table S1, Supplementary Material online).
Table 1.
Number of Polymorphic and Fixed Nucleotide Differences between AT and GC in Drosophila melanogaster Raleigh Sample
| Type of Mutation | AT to GC |
GC to AT |
GC-to-AT/AT-to-GC | |||
|---|---|---|---|---|---|---|
| No. of Nucleotide Differences | Percentage of Deviation | No. of Nucleotide Differences | Percentage of Deviation | |||
| Autosomal | ||||||
| Noncoding | Fixed | 338,495 | +11.6 | 430,988 | −7.6 | 1.27 |
| Polymorphic | 240,762 | −12.8 | 459,802 | +8.3 | 1.91 | |
| Synonymous | Fixed | 73,122 | +27.5 | 101,769 | −13.4 | 1.39 |
| Polymorphic | 12,766 | −55.2 | 74,191 | +27.0 | 5.81 | |
| X linked | ||||||
| Noncoding | Fixed | 71,330 | +5.9 | 92,009 | −4.1 | 1.28 |
| Polymorphic | 24,249 | −14.1 | 44,210 | +9.9 | 1.82 | |
| Synonymous | Fixed | 8,167 | +21.2 | 16,071 | −8.2 | 1.97 |
| Polymorphic | 841 | −62.9 | 7,326 | +24.2 | 8.71 | |
Consistent with previous reports, the ratios of AT and GC fixed and polymorphic variants (table 1) show that derived AT variants are more common than are derived GC variants (relative to fixations). Although this bias was observed across all site types, the magnitude was heterogeneous (table 1). For synonymous mutations, the deviation from expectation ranged from 8.2% to 62.9%, but the level of deviation was only 10% for noncoding variants. For noncoding variants, the ratios of AT to GC fixations on the autosomes and X chromosome were very similar (A: 1.28X vs. X: 1.29X), whereas the ratio of AT-to-GC polymorphisms was slightly higher on the autosomes than on the X chromosome (A: 1.91X vs. X: 1.82X). A comparable analysis of synonymous sites revealed three patterns. First, as noted before, the magnitude of the fixation bias for AT mutations relative to GC mutations was greater than that for noncoding DNA. Second, the AT fixation bias was considerably greater on the X chromosome (1.97X) than on the autosomes (1.39X). Finally, compared with noncoding DNA, synonymous AT variants showed a dramatic excess of polymorphisms (5.81X for autosomes and 8.71X for X chromosome). All these patterns were also observed for a small sample of Malawi D. melanogaster genomes (supplementary table S1, Supplementary Material online) and for all codon families (supplementary table S2, Supplementary Material online).
Frequency Distribution
The observations that AT variants are fixing at a higher rate than GC mutations at synonymous sites and that the ratio of AT-to-GC fixations is greater in regions of higher crossing-over support the idea that synonymous AT mutations are weakly favored by natural selection (Bauer DuMont et al. 2009), in contrast to standard models of mutation–selection–drift equilibrium of codon bias in D. melanogaster, which posits that GC variants are favored. However, the dramatic excess of AT polymorphisms at synonymous sites supports the standard model in that weakly deleterious mutations are expected to appear in excess as polymorphisms (e.g., Kimura 1983; Ohta and Gillespie 1996). Alternatively, an excess of AT synonymous polymorphisms could also be explained as a recent change in mutation bias. One approach for distinguishing between these hypotheses is through analysis of the frequency spectrum. The standard mutation–selection–drift model predicts that AT polymorphisms should show greater skew toward rare alleles than GC polymorphisms and that this difference should be greater for genes experiencing greater selection for codon bias.
The allele frequency spectra for AT and GC mutations are shown in supplementary figure S2, Supplementary Material online. The proportion of synonymous AT mutations that are singletons is 23.2%, which is lower than the 24.3% for GC mutations. However, for noncoding AT mutations, the proportion of singletons is higher than that of noncoding GC mutations (35.2% vs. 32.8%). Tajima’s D and Fu and Li’s D* were used to summarize the skew of the site frequency spectrum for unpolarized (i.e., folded) and polarized (i.e., unfolded) data, respectively (table 2). Neither test statistic differed from the neutral expectation generated from simulated data (data not shown). Nevertheless, the MWU test of frequency spectra revealed several general patterns. First, noncoding mutations show greater skew toward low frequency variants than do synonymous mutations. Second, X-linked polymorphisms also show greater skew toward low-frequency variants than do autosomal polymorphisms. Third, and perhaps most surprisingly, Tajima’s D test statistic for synonymous sites at which the derived variant is an A or T shows a greater positive value than do comparable sites at which the derived state is G or C. This third pattern is consistent with the hypothesis that AT polymorphisms are associated with higher fitness than GC polymorphisms. To further investigate this pattern, we characterized the frequency spectrum specifically for derived variants using Fu and Li’s D* and found the same pattern—derived synonymous AT variants show less skew toward rare alleles than do derived synonymous GC variants (table 2). The different pattern of the frequency spectrum of AT versus GC polymorphisms is particularly pronounced for the X chromosome. Because of the reference sequence bias inherent to the sequencing technology, these analyses were repeated for sites with more stringent quality cutoff of Q40. These more stringent standards reduce the coverage and thus the power, but the observed patterns remain (supplementary table S3, Supplementary Material online).
Table 2.
Summary Statistics of the Frequency Spectra for AT and GC Mutations
| Type of Mutation |
Region | Tajima’s D |
Fu and Li’s D* |
P Value of MWU Test | ||
|---|---|---|---|---|---|---|
| AT to GC | GC to AT | AT to GC | GC to AT | |||
| Noncoding | Autosome | −0.625 | −0.698 | −0.632 | −0.828 | 0.0002119 |
| X | −0.658 | −0.734 | −0.742 | −0.969 | 7.424e−05 | |
| Synonymous | Autosome | −0.109 | −0.003 | 0.139 | 0.166 | 8.946e−11 |
| X | −0.456 | −0.227 | −0.959 | 0.004 | 1.519e−11 | |
| High crossing-over rate | −0.063 | 0.002 | 0.247 | 0.243 | 1.564e−10 | |
| Low crossing-over rate | −0.016 | −0.019 | 0.166 | 0.079 | 6.651e−11 | |
| Highly expressed genes | −0.324 | −0.096 | −0.446 | −0.059 | 6.456e−11 | |
| Lowly expressed genes | −0.182 | 0.055 | 0.064 | 0.286 | 1.256e−10 | |
Multiple lines of evidence point to a strong positive correlation between gene expression and selection on codon bias (Akashi and Eyre-Walker 1998). If selection contributes to the different frequency spectra for AT versus GC mutations, then genes under stronger selection for codon bias should show a greater difference in GC versus AT frequency. Supporting this prediction, we found that highly expressed genes are much more strongly skewed toward low-frequency GC polymorphisms compared with AT polymorphisms (table 2). Similarly, lowly expressed genes went in the same direction though the magnitude of the difference between AT and GC polymorphisms was much smaller.
Under weak selection, the differences between AT and GC polymorphisms should be greater in regions of higher recombination (Hill and Robertson 1966). Because the number of polymorphic sites for the genes in the no-crossing-over (N) category is too small to conduct statistical tests on allele frequency spectra, we combined the no-crossing over genes with those from the low crossing-over category (L) and compared these genes (0–2.93 cM/Mb) with those from the high-crossing-over category (>2.93 cM/Mb). The site frequency spectra corresponding to the genes with low crossing-over are not significantly different from noncoding polymorphisms (Kolmogorov–Smirnov test, P = 0.8253, table 2). For synonymous sites, Tajima’s D supports the idea that sites segregating a derived AT are less skewed toward rare alleles in regions of higher crossing over, whereas Fu and Li’s D* rejects the hypothesis. Overall, there is no strong support for an effect of recombination on the frequencies of derived AT versus GC polymorphisms.
To investigate the frequency spectra in more detail, we divided the genes into four groups: highly expressed and high recombination; lowly expressed and high recombination; highly expressed and low recombination; and lowly expressed and low recombination (table 3). The rationale was that highly expressed genes should experience stronger selection on codon usage and that the efficacy of selection on codon usage should also be greater in regions of higher recombination. In general, we found that expression level played a more important role than recombination rate on codon usage. For Tajima’s D, GC polymorphisms are more skewed toward rare alleles than AT polymorphisms in three of the groups we compared. The only exception was in the lowly expressed/low-recombination genes, where selection was expected to be least effective. Similar results were obtained for Tajima’s Fu and Li’s D*.
Table 3.
Summary Statistics of the Frequency Spectra for AT and GC Mutations under Different Expression and Recombination Criteria
| Highly Expressed |
Lowly Expressed |
|||||||
|---|---|---|---|---|---|---|---|---|
| AT to GC |
GC to AT |
AT to GC |
GC to AT |
|||||
| Da | D*b | Da | D*b | Da | D*b | Da | D*b | |
| High crossing-over rate | −0.1326 | 0.0730 | −0.0388 | 0.1596 | −0.0398 | 0.2231 | 0.0496 | 0.3152 |
| Low crossing-over rate | −0.3726 | −0.3993 | −0.0250 | 0.0938 | 0.1966 | 0.4605 | 0.0473 | 0.1246 |
aTajima’s D.
bFu and Li’s D*.
Discussion
Previous analyses of D. melanogaster divergence suggested the possibility that AT mutations may be favored in this species (Holloway et al. 2008; Bauer DuMont et al. 2009). Our analysis of divergence supports the conclusion of Bauer Dumont et al. (2009) that AT-biased substitution is positively correlated with recombination. Additionally, we found that the AT-bias substitution rate is positively correlated with ancestral GC content in the D. melanogaster lineage. These results cannot be explained simply by biased gene conversion (Galtier et al. 2006) but are consistent with the idea that ancestrally GC-rich regions departs farther from equilibrium base composition than are ancestrally AT-rich regions.
One interpretation of these divergence patterns is that there has been a shift of codon preference, such that AT-ending codons are on average beneficial (whereas GC-ending codons were ancestrally favored). If this were the case then AT codons should occur at higher frequencies than GC codons, and this difference should be magnified in regions of higher recombination as a result of reduced Hill–Robertson effects. Our results generally support these population genetic predictions, though it is worth noting the effects are small, and that the effect of recombination rate variation on the frequency spectrum of AT versus GC polymorphisms was equivocal. Genes showing higher recombination showed a more negative Tajima’s D for GC than for AT sites, whereas no such difference was observed for Fu and Li’s D*. However, considering only genes that are highly expressed and are located in high recombination regions, both Tajima’s D and Fu and Li’s D* show that GC polymorphisms are more skewed toward rare alleles than AT polymorphisms (table 3). This result appears to be robust to quality and coverage variation.
Under an alternative hypothesis that the excess AT polymorphisms in D. melanogaster result from a recent relaxation of selection, one predicts that genes that were under the strongest selection ancestrally would show the greatest relative accumulation of AT polymorphisms. However, it is difficult to understand why under this model AT mutations would occur at higher frequency on average, than GC mutations. If AT mutations are segregating at higher frequency due to alignment errors, we would expect the pattern to be stronger in noncoding than coding regions. However, the patterns are much stronger for synonymous than for noncoding sites. In contrast to our results, Zeng and Charlesworth (2009) found in a Zimbabwe sample of D. melanogaster that AT polymorphisms had lower frequencies than GC polymorphisms. Our analysis included a much larger number of sites and alleles, though we cannot rule out that the patterns of base composition polymorphism are different in the Zimbabwe and Raleigh populations. Additionally, given the positive correlation of AT-bias substitution rate and GC content, the observation of positive correlation between AT substitution bias and recombination rate can be explained by relaxation of selection only if highly expressed genes are clustered in the regions of high recombination rate. However, expression level and recombination rate are not positively correlated (Spearman’s ρ = 0.01500984, P value = 0.1877). In summary, though it is surprising that natural selection could favor a shift in codon preference, it appears to be the most parsimonious explanation for population genetic patterns of polymorphism and divergence in D. melanogaster.
Preferred codons are thought to correspond to transfer RNA (tRNA) abundance (Ikemura 1985; Dong et al. 1996; Shah and Gilchrist 2010; Supek et al. 2010). A switch of preferred codon in yeast has been observed for species that have undergone whole genome duplication. Copy number of tRNA genes evolved differentially and led to a switch of the preferred codons (Lin et al. 2006). However, the turnover of tRNA genes does not appear to be directly coupled with the shift of codon usage in Drosophila because there is no anticodon change on D. willistoni branch where the pattern of codon usage bias was shifted (Rogers et al. 2010). Because most of the tRNA genes are conserved between D. melanogaster and D. simulans, the codon shift model makes a strong prediction that the expression profile of tRNA has diverged between the two species to accommodate the shift of codon preference in D. melanogaster.
Supplementary Material
Supplementary tables S1–S4 and figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Grace Yuh Chwen Lee and Kristian Stevens for providing D. melanogaster, D. simulans, and D. yakuba sequence alignments and conservative gene sets among those species. This work was supported by grants NSC 9402917-1-007–011 to Y.-P.P., NSC 97-2621-B-002-004-MY3 to C.-T.T., and NIH HG02942 to C.H.L.
Literature Cited
- Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics. 1996;144:1297–1307. doi: 10.1093/genetics/144.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Distinguishing the effects of mutational biases and natural selection on DNA sequence variation. Genetics. 1997;147:1989–1991. doi: 10.1093/genetics/147.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Eyre-Walker A. Translational selection and molecular evolution. Curr Opin Genet Dev. 1998;8:688–693. doi: 10.1016/s0959-437x(98)80038-5. [DOI] [PubMed] [Google Scholar]
- Akashi H, Goel P, John A. Ancestral inference and the study of codon bias evolution: implications for molecular evolutionary analyses of the Drosophila melanogaster subgroup. PLoS One. 2007;2:e1065. doi: 10.1371/journal.pone.0001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Kliman RM, Eyre-Walker A. Mutation pressure, natural selection, and the evolution of base composition in Drosophila. Genetica. 1998;102–103:49–60. [PubMed] [Google Scholar]
- Akashi H, Schaeffer SW. Natural selection and the frequency distribution of “silent” DNA polymorphism in Drosophila. Genetics. 1997;146:195–307. doi: 10.1093/genetics/146.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, et al. Molecular evolution in the Drosophila melanogaster species subgroup: frequent parameter fluctuations on the timescale of molecular divergence. Genetics. 2006;172:1711–1726. doi: 10.1534/genetics.105.049676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DuMont V, Fay JC, Calabrese PP, Aquadro CF. DNA variability and divergence at the notch locus in Drosophila melanogaster and D. simulans: a case of accelerated synonymous site divergence. Genetics. 2004;167:171–185. doi: 10.1534/genetics.167.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DuMont VL, Singh ND, Wright MH, Aquadro CF. Locus-specific decoupling of base composition evolution at synonymous sites and introns along the Drosophila melanogaster and Drosophila sechellia lineages. Genome Biol Evol. 2009;1:67–74. doi: 10.1093/gbe/evp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ. The frequency distribution of nucleotide variation in Drosophila simulans. Mol Biol Evol. 2001;18:1343–1352. doi: 10.1093/oxfordjournals.molbev.a003918. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Whitley P. Molecular population genetics of Xdh and the evolution of base composition in Drosophila. Genetics. 2002;162:1725–1735. doi: 10.1093/genetics/162.4.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini DB. Experimental reduction of codon bias in the Drosophila alcohol dehydrogenase gene results in decreased ethanol tolerance of adult flies. J Evol Biol. 2004;17:779–785. doi: 10.1111/j.1420-9101.2004.00725.x. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M, Aguade M. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev. 2002;12:640–649. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiston-Lavier AS, Singh ND, Lipatov M, Petrov DA. Drosophila melanogaster recombination rate calculator. Gene. 2010;463:18–20. doi: 10.1016/j.gene.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Bazin E, Bierne N. GC-biased segregation of noncoding polymorphisms in Drosophila. Genetics. 2006;172:221–228. doi: 10.1534/genetics.105.046524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Charlesworth B. Non-neutral processes drive the nucleotide composition of non-coding sequences in Drosophila. Biol Lett. 2008;4:438–441. doi: 10.1098/rsbl.2008.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Ponting CP. Variable strength of translational selection among 12 Drosophila species. Genetics. 2007;177:1337–1348. doi: 10.1534/genetics.107.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense W, et al. Experimentally increased codon bias in the Drosophila Adh gene leads to an increase in larval, but not adult, alcohol dehydrogenase activity. Genetics. 2010;184:547–555. doi: 10.1534/genetics.109.111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Holloway AK, Begun DJ, Siepel A, Pollard KS. Accelerated sequence divergence of conserved genomic elements in Drosophila melanogaster. Genome Res. 2008;18:1592–1601. doi: 10.1101/gr.077131.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Gene genealogies and the coalescent process. In: Futuyma D, Antonovics J, editors. Oxford surveys in evolutionary biology. Oxford (United Kingdom): Oxford University Press; 1990. pp. 1–44. [Google Scholar]
- Hudson RR. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Keightley PD, et al. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19:1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern AD, Begun DJ. Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes. Mol Biol Evol. 2005;22:51–62. doi: 10.1093/molbev/msh269. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge (United Kingdom): Cambridge University Press; 1983. [Google Scholar]
- Langley CH, et al. Genomic variation in natural populations of Drosophila melanogaster. Genetics. 2012;192:533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Byrnes JK, Hwang JK, Li WH. Codon-usage bias versus gene conversion in the evolution of yeast duplicate genes. Proc Natl Acad Sci U S A. 2006;103:14412–14416. doi: 10.1073/pnas.0606348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Mouchiroud D, Duret L. Neutral effect of recombination on base composition in Drosophila. Genet Res. 2003;81:79–87. doi: 10.1017/s0016672302006079. [DOI] [PubMed] [Google Scholar]
- McVean GA, Vieira J. The evolution of codon preferences in Drosophila: a maximum-likelihood approach to parameter estimation and hypothesis testing. J Mol Evol. 1999;49:63–75. doi: 10.1007/pl00006535. [DOI] [PubMed] [Google Scholar]
- McVean GA, Vieira J. Inferring parameters of mutation, selection, and demography from patterns of synonymous site evolution in Drosophila. Genetics. 2001;157:245–257. doi: 10.1093/genetics/157.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama EN, Hartl DL. Codon usage bias and base composition of nuclear genes in Drosophila. Genetics. 1993;134:847–858. doi: 10.1093/genetics/134.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bauer DuMont VL, Hubisz MJ, Aquadro CF. Maximum likelihood estimation of ancestral codon usage bias parameters in Drosophila. Mol Biol Evol. 2007;24:228–235. doi: 10.1093/molbev/msl146. [DOI] [PubMed] [Google Scholar]
- Nolte V, Schlotterer C. African Drosophila melanogaster and D. simulans populations have similar levels of sequence variability, suggesting comparable effective population sizes. Genetics. 2008;178:405–412. doi: 10.1534/genetics.107.080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Gillespie JH. Development of neutral and nearly neutral theories. Theor Popul Biol. 1996;49:128–142. doi: 10.1006/tpbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala FJ. Switch in codon bias and increased rates of amino acid substitution in the Drosophila saltans species group. Genetics. 1999;153:339–350. doi: 10.1093/genetics/153.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HH, Bergman CM, Griffiths-Jones S. The evolution of tRNA genes in Drosophila. Genome Biol Evol. 2010;2:467–477. doi: 10.1093/gbe/evq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Gilchrist MA. Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLoS Genet. 2010;6:e1001128. doi: 10.1371/journal.pgen.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Shields DC, Sharp PM, Higgins DG, Wright F. “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- Singh ND, Arndt PF, Petrov DA. Genomic heterogeneity of background substitutional patterns in Drosophila melanogaster. Genetics. 2005;169:709–722. doi: 10.1534/genetics.104.032250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Arndt PF, Petrov DA. Minor shift in background substitutional patterns in the Drosophila saltans and willistoni lineages is insufficient to explain GC content of coding sequences. BMC Biol. 2006;4:37. doi: 10.1186/1741-7007-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Bauer DuMont VL, Hubisz MJ, Nielsen R, Aquadro CF. Patterns of mutation and selection at synonymous sites in Drosophila. Mol Biol Evol. 2007;24:2687–2697. doi: 10.1093/molbev/msm196. [DOI] [PubMed] [Google Scholar]
- Stenico M, Lloyd AT, Sharp PM. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 1994;22:2437–2446. doi: 10.1093/nar/22.13.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol. 2007;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- Supek F, Skunca N, Repar J, Vlahovicek K, Smuc T. Translational selection is ubiquitous in prokaryotes. PLoS Genet. 2010;6:e1001004. doi: 10.1371/journal.pgen.1001004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Shimizu T. Local changes in GC/AT substitution biases and in crossover frequencies on Drosophila chromosomes. Mol Biol Evol. 2001;18:606–619. doi: 10.1093/oxfordjournals.molbev.a003841. [DOI] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. Estimating selection intensity on synonymous codon usage in a nonequilibrium population. Genetics. 2009;183:651–662, 651SI–623SI. doi: 10.1534/genetics.109.101782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. Studying patterns of recent evolution at synonymous sites and intronic sites in Drosophila melanogaster. J Mol Evol. 2010;70:116–128. doi: 10.1007/s00239-009-9314-6. [DOI] [PubMed] [Google Scholar]