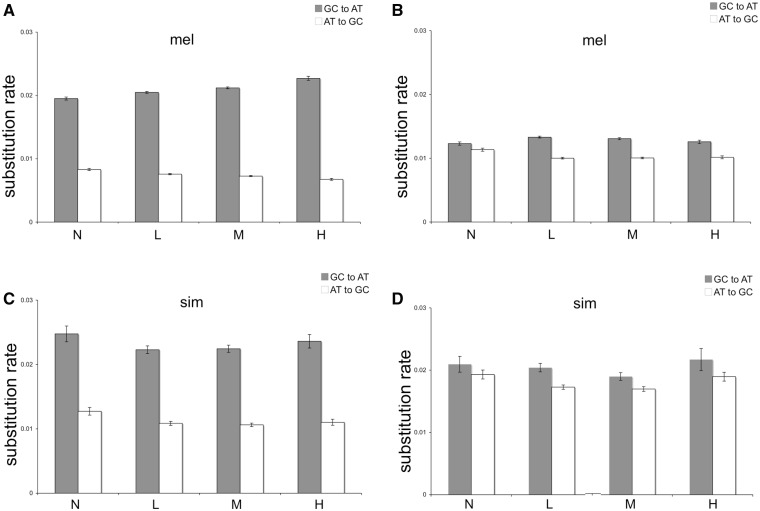
Fig. 3.—
Relationship between recombination rate and the profile of base composition evolution. Mean substitution rate (±Standard Error) of AT and GC nucleotide substitutions of Drosophila melanogaster (A: exon and B: intron) and D. simulans genome (C: exon and D: intron). Within each data set, the two types of substitutions were subdivided into categories according to frequency of crossing over: high (H), >3.9 cM/Mb; medium (M), 2.93–3.9 cM/Mb; low (L), 0.27–2.93 cM/Mb; and none (N), <0.27.