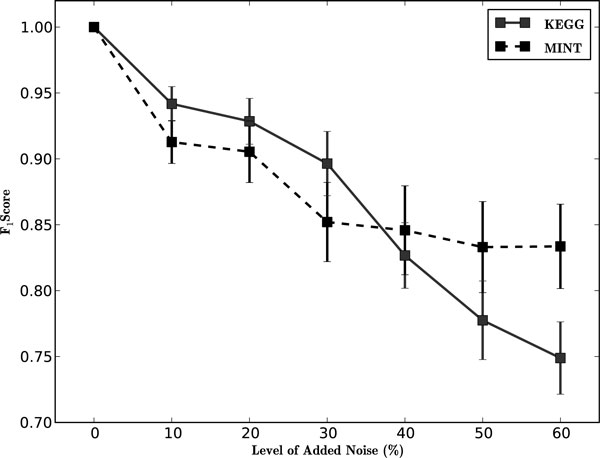
Figure 4.
Recovering functional modules contaminated with noise. The original gene lists for KEGG and MINT (N = 22 and N = 24 respectively) were compiled and increasing levels of noise were added to each set, where each was then considered a new list. Spectral clustering was run on each gene list and statistical significance based on functional coherence was determined for all the the underlying modules. Statistically significant modules were assembled and together they made up the positively labeled genes. Shown here are the averaged results of the gene sets, with standard error bars at each level of noise.