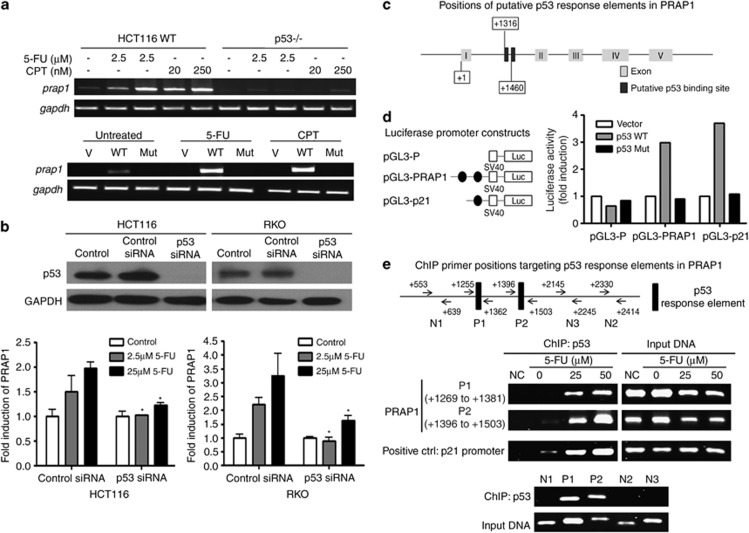
Figure 2.
Induction of PRAP1 by DNA-damaging agents is dependent on p53. (a, top panel) Cells without p53 failed to induce PRAP1 when treated with DNA-damaging agents. Reverse transcriptase (RT)-PCR gel picture showing the expression of prap1 and gapdh (glyceraldehyde 3-phosphate dehydrogenase) in HCT116 cells and significantly reduced expression of prap1in the p53-nulll HCT116 derivative, p53−/−, after treatment with the indicated doses of 5-FU and CPT for 24 h. (a, bottom panel) Reintroduction of wild-type p53 into p53−/− cells restored PRAP1 induction by DNA-damaging agents. RT-PCR gel picture showing the expression of prap1 and gapdh in p53−/− cells after transfection with either empty vector control (V) or wild-type p53 (WT) or mutant p53 (Mut), followed by treatment with either 5-FU (25 μM) or CPT (20 nM) for 24 h. Transfection with WT restored PRAP1 induction by 5-FU and CPT. (b, top panel) Western blot picture showing p53 protein was efficiently knocked down by siRNA in both HCT116 and RKO cells. (b, bottom panel) Both p53-depleted cell lines failed to induce PRAP1 upon 5-FU treatment. Graphs showing induction of prap1 expression in HCT116 and RKO cells after treatment with 5-FU (2.5 or 25 μM) for 24 h. Real-time RT-PCR was performed and relative expression of prap1 were normalized against gapdh and calculated as fold induction. p53 knockdown cells showed marked reduction in the induction of PRAP1 expression after 5-FU treatment as compared with the respectively control siRNA-treated cells (*P<0.05). (c) Schematic diagram of p53 binding sites in PRAP1 gene construct. Two p53 binding sites were identified in intron 1 of the PRAP1 gene, starting at +1316 and +1460, while the transcription start site of PRAP1 gene is denoted as +1. (d, left panel) The two p53-response elements (blue) within the first intron 1 of PRAP1 gene were cloned into the luciferase reporter plasmid (pGL3-Promoter) upstream of the SV 40 promoter to generate the pGL3-PRAP plasmid. (d, right panel) Figure showing the fold induction of SV40 promoter activity of pGL3-P, pGL3-PRAP and pGL3-p21 cotransfected with pcDNA vector, pcDNA-p53 and pcDNA-p53Mut in p53−/− cells for 24 h. For each transfection, the Firefly luciferase activity was normalized with the Renilla reniformis luciferase activity by the cotransfected pRL-TK (thymidine kinase promoter-Renilla luciferase reporter plasmid). The relative activity of each construct is compared against the activity of the pGL3-P. pGL3-P, basic luciferase promoter; pGL3-PRAP, basic luciferase promoter plasmid constructed with the two p53 binding elements of PRAP1 gene; pGL3-p21, basic luciferase promoter plasmid constructed with the p53 binding elements of p21 gene (serving as a positive control for comparison); Vector: pcDNA vector; p53 WT: pcDNA with wild-type p53 construct; p53 Mut: pcDNA with mutant p53. (e, top panel) Schematic diagram of chromatin immunoprecipitation (ChIP) primers used to amplify the two p53-response elements and non-specific regions in intron 1 of PRAP1 gene. (e, middle panel) p53 directly interacted with the two p53-response elements in PRAP1 gene. HCT116 cells were treated with 5-FU (25 or 50 μM) for 24 h. The chromatin bound with p53 were isolated and amplified by PCR. A primer pair recognizing the known p53-response element on p21 promoter was used as a positive control. (e, bottom panel) p53 specifically bound to the p53-response elements (P1, P2). Primer pairs that align to non-specific regions in intron 1 did not amplify a band (N1, N2 and N3). The ChIP-precipitated DNA and input DNA were from cells treated with 25 μM 5-FU