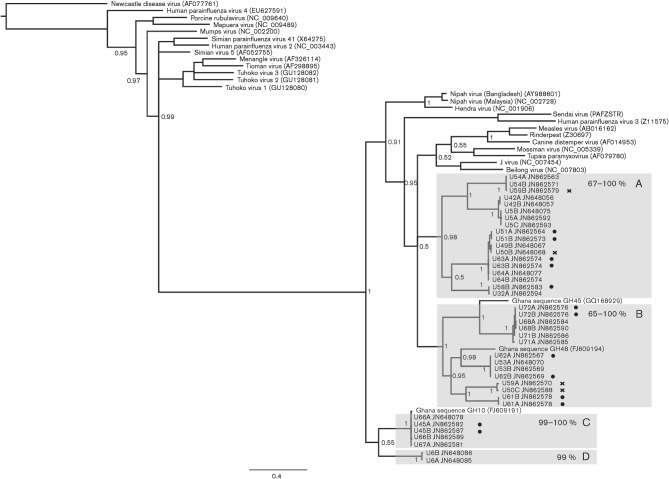
Fig. 2.
Diversity of paramyxoviruses in pooled E. helvum urine samples detected by using RMH-targeted PCR. Phylogenetic tree for a 439 bp gap-stripped alignment of the polymerase gene of members of the subfamily Paramyxovirinae, including sequences generated in this study and publicly available paramyxovirus genome sequences (as per Fig. 1) and partial paramyxovirus sequences available from this species (Ghanaian sequences GH45, GH48 and GH10). Relevant posterior probability values are shown. Bar, 0.4 expected nucleotide substitutions per site. Individual sample IDs are followed by letters denoting the clone and the GenBank accession for the sequence. Groups containing previously uncharacterized sequences that display a common phylogenetic origin supported by high posterior probability values (≥0.95) are highlighted by lettered grey boxes. Adjacent to each box letter is the range of nucleotide identities among sequences in the clade. Clones derived from samples that contained sequences belonging to more than one of these phylogenetically distinct clades were marked with a cross. Sequences derived from samples that were positive only with this PCR (and negative with PMV-PCR) are marked with a dot.