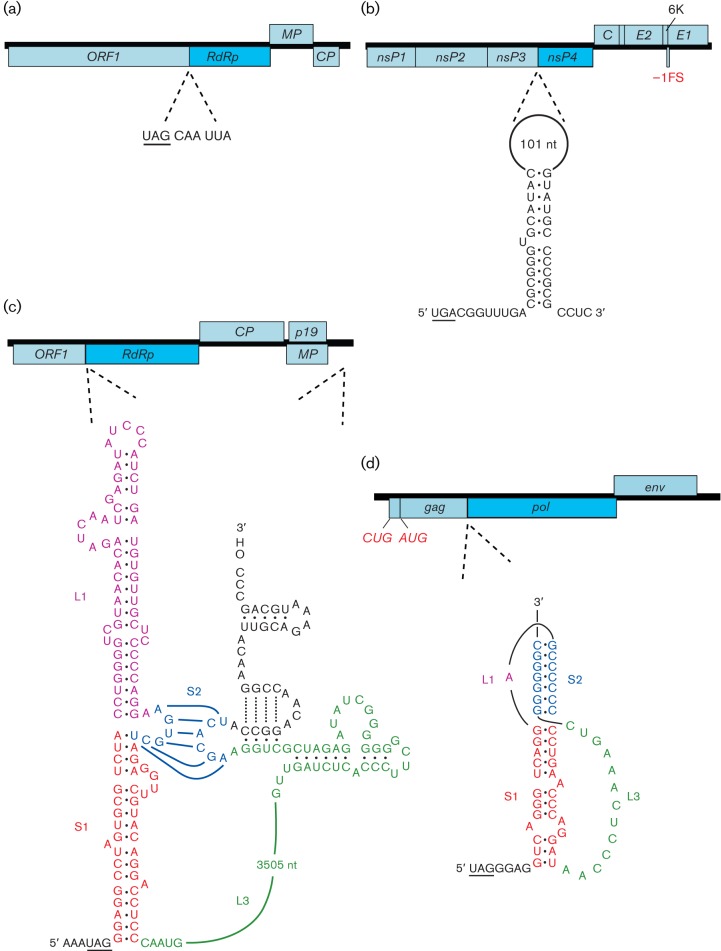
Fig. 7.
Programmed stop codon readthrough in viral mRNAs. In each of the four examples, the genome is indicated as an ORF map, with the location of the readthrough site shown by dotted lines. In tobacco mosaic tobamovirus (a), only a short local sequence context 3′ of the recoded UAG is required for efficient readthrough. In Venezuelan equine encephalitis alphavirus (b), carnation italian ringspot tombusvirus (c) and murine leukemia gammaretrovirus (d), the 3′ stimulator is an RNA secondary structure: an extended stem–loop in (b), an RNA pseudoknot in (d) and long-range base pairing in (c). Spliced and subgenomic RNAs are not shown and polyprotein cleavage products are only indicated where specifically relevant. ‘FS’ indicates a ribosomal frameshift site.