Fig. 2.
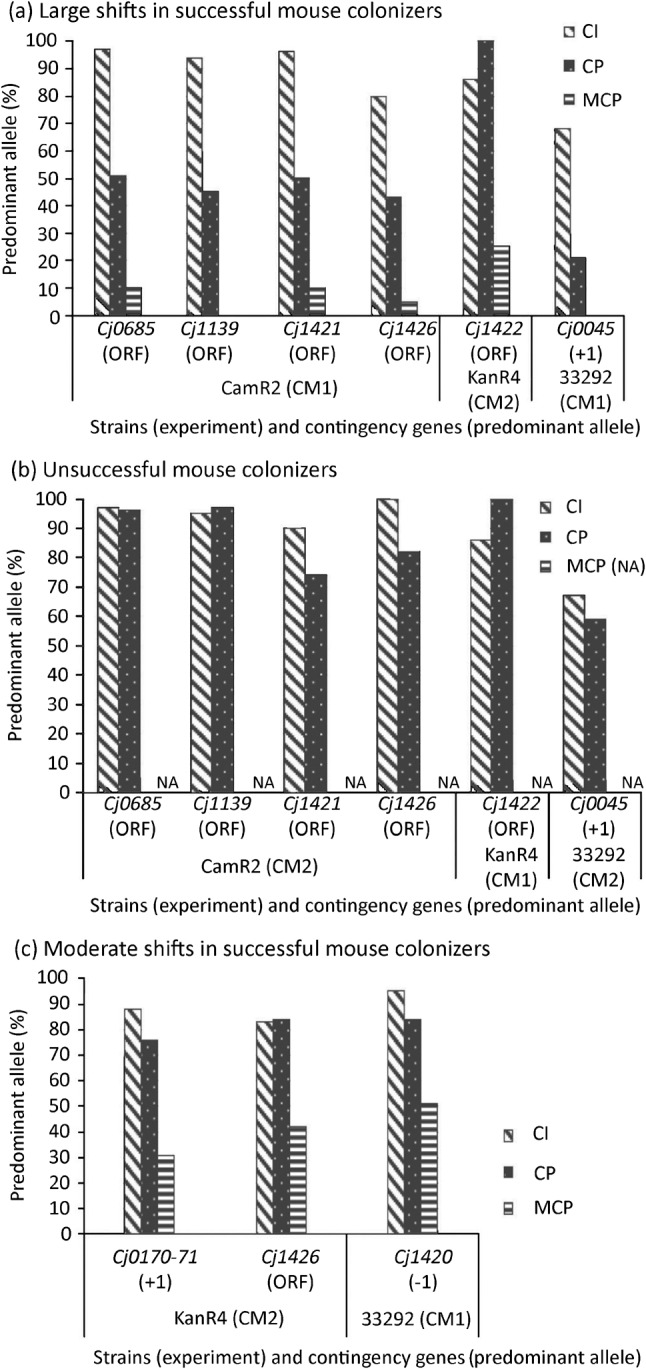
Frequency (percentage) of the predominant allele in C. jejuni populations. The initial predominant allele is defined as the highest-frequency allele in a non-passaged C. jejuni (CI) population grown in culture. Contingency genes were amplified by PCR, and fragment and/or sequence analyses were used to determine allele frequencies in a non-passaged population (CI), after chicken passage (CP), or after passage through chickens and then mice (MCP). (a) ‘Successful mouse colonizers’ colonized mice at high frequency after passage through chickens. Predominant allele frequencies in CI and CP populations were significantly different (P<0.05), except for Cj1422 in KanR4 populations in CM2 (P = 0.23). Predominant allele frequencies in CP and MCP populations also were significantly different (P<0.05). (b) ‘Unsuccessful mouse colonizers’ did not colonize mice at high frequency after chicken passage. We did not observe significant differences in predominant allele frequencies between CI and CP populations (P>0.05), except for Cj1426 in the CamR2 population (CM2) (P = 3.3×10−6). MCP populations were not available (na) because mice were not colonized. With the exception of locus Cj1422, allele frequencies for all contingency loci in successful CP populations shown in (a) were significantly different from unsuccessful CP populations shown in (b) (P<0.05). (c) MCP populations experienced moderate allele shifts after passage through chickens and then mice. Allele frequencies for contingency loci in CI and MCP populations were significantly different (P<0.05). Allele frequencies for contingency loci in CI and CP populations were significantly different (P<0.05), except for Cj1426 in KanR4 populations (CM2). For alleles predicted to encode a complete ORF, see Table 1.
