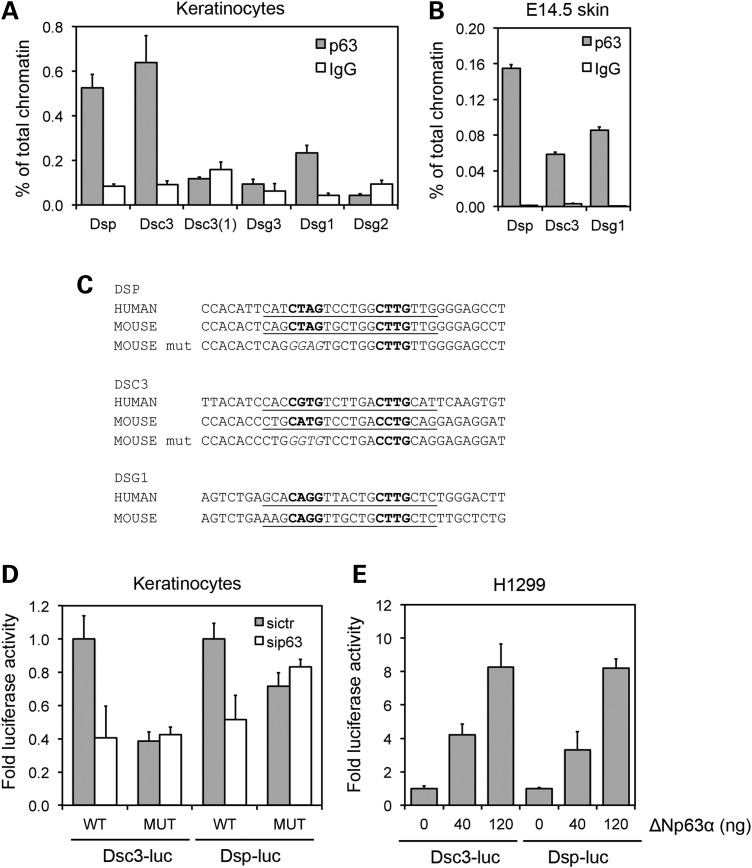
Figure 5.
Identification of desmosomal genes directly controlled by p63. (A) ChIP-qPCR of mouse keratinocyte chromatin was performed using p63 polyclonal antibodies (H-137) (grey bars) and rabbit IgG (white bars) as negative control. Strong p63 binding was observed in mouse genomic regions corresponding to the human p63-binding regions in Dsp and Dsc3. Binding was observed also in a genomic region upstream of Dsg1. Data are representative of three independent experiments. Error bars denote SEM. (B) ChIP-qPCR of E14.5 skin chromatin showing that p63 efficiently bound Dsp, Dsc3 and Dsg1 regulatory regions in vivo. Data are represented as in (A). (C) Sequence of the conserved p63-binding motif in human and mouse in the Dsp, Dsc3 and Dsg1 regulatory regions is underlined. The core consensus is in bold. p63-binding site in the regulatory regions was identified by MatInspector (http://www.genomatix.de/). Two nucleotides in the core sequence were mutated in the Dsp and Dsc3 core sequence as indicated (p63 mut) for the luciferase assay. (D) Luciferase activity of the Dsp and Dsc3 regulatory regions cloned in the pGL3-Luc vector (WT) is strongly inhibited in p63 knockdown (white bars) when compared with control mouse keratinocytes (grey bars). In contrast, Dsp and Dsc3 regulatory regions with a mutant p63-binding site as in (C) (MUT) were no longer responsive to p63 knockdown. Data are expressed relative to the WT p63-binding site treated with the siRNA control (siCTR). (E) The luciferase assay in H1299 cells transfected with the indicated concentrations of ΔNp63alpha expressing vector and Dsp and Dsc3 luciferase reporters. Data are representative of three independent experiments. Error bars denote SEM.