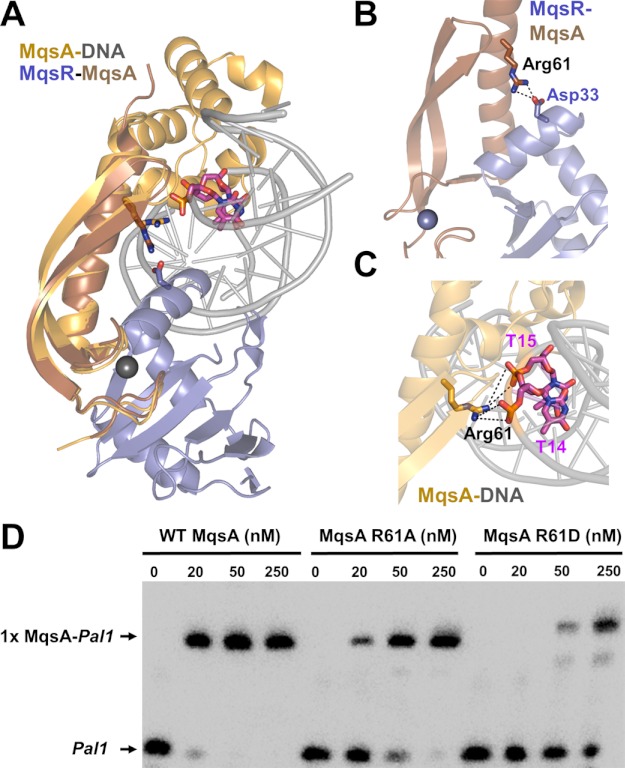
FIGURE 5.
MqsR and DNA have overlapping binding sites on MqsA. A, model of the hypothetical MqsR-MqsA-PmqsRA ternary complex. The model was generated by superimposing the MqsR-MqsA-N crystal structure (PDB 3HI2; colored blue/brown, respectively) with the MqsA-DNA crystal structure (PDB 3O9X; colored orange/gray, respectively) using the respective MqsA N-terminal domains; for clarity, only one monomer of MqsA from the MqsA-DNA crystal structure is shown. As modeled, there are structural clashes between MqsR and the DNA. The MqsA binding sites for MqsR and the PmqsRA DNA partially overlap as MqsA residue Arg-61 provides electrostatic interactions in both the toxin- and DNA-bound states; Arg-61 from both structures shown as sticks. Asp-33 (MqsR, blue) and the phosphate backbone from nucleotides thymine 14 and 15 (pink) are also shown; nitrogen atoms in dark blue, oxygen atoms in red. B, zoom-in view of overlay in A of just the MqsR-MqsA-N structure. MqsA residue Arg-61 forms a salt bridge with MqsR Asp-33. C, zoom-in view of overlay in A of just the MqsA-DNA structure. MqsA residue Arg-61 forms a salt bridge with the phosphate backbone of nucleotides thymine 14 and 15 from one strand. Electrostatic interactions in B and C are indicated by black dashed lines. D, EMSA with increasing amounts of WT MqsA (lanes 1–4), MqsA R61A (lanes 5–8), or MqsA R61D (lanes 9–12) incubated with biotin-labeled palindrome 1 (Pal1) of PmqsRA.