FIGURE 1.
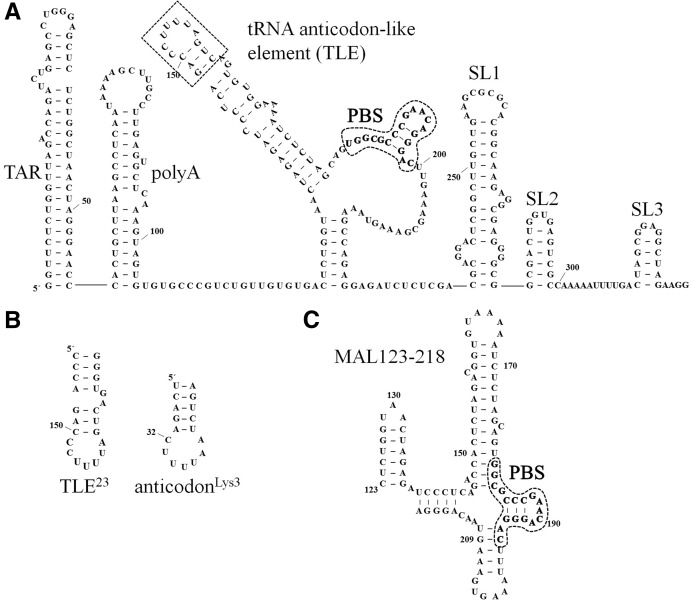
(A) Predicted secondary structure of a 329-nt RNA derived from the HIV-1 NL4-3 5′ UTR with the TLE sequence boxed and PBS circled and in bold. Although only the TLE loop sequence resembles the anti-codon domain of tRNALys3, we have included the flanking 3-bp stem domain in our definition of the TLE to emphasize the presentation of anti-codon-like nucelotide in an exposed loop. The NL4-3 “PBS/TLE” RNA used in this study (105 nt) contains nt 125–223 plus three additional 5′-G-C-3′ pairs to stabilize the terminal RNA stem and aid in vitro transcription. The 330-nt 5′ UTR RNA used in this study contains the 329-nt sequence shown with an additional 5′-G residue for in vitro transcription. The ΔTARpolyA mutation is a deletion of nt 1–124, the ΔSL3 mutation is a deletion of nt 300–329, and the ΔSL2,3 mutation is a deletion of nt 284–329. (B) The sequence of a 23-mer derived from the vRNA (TLE23) is shown next to the anti-codon stem–loop of tRNALys3 for comparison. This construct contains two additional nonviral 5′-C-G-3′ pairs to stabilize the terminal RNA stem. (C) Predicted secondary structure of the MAL construct derived from nt 123–218 of the HIV-1 MAL isolate is shown, with the PBS sequence circled and in bold. The A-rich stem–loop examined in Puglisi and Puglisi (1998) includes nt 156–172.