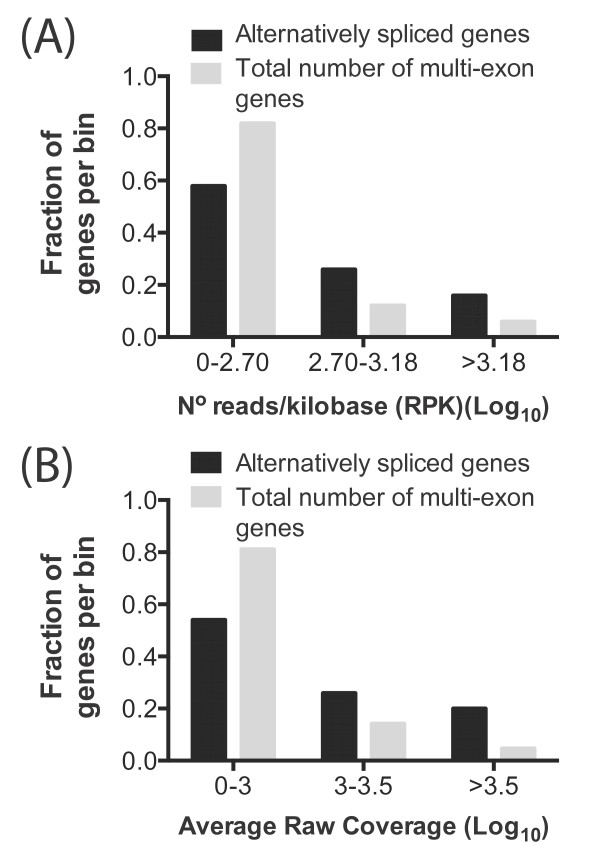
Figure 5.
The ability to detect alternatively spliced isoforms using RNA-seq data is dependent on read coverage. Shown are fraction of alternatively spliced (from a total of 50) (Black bars) and multi-exonic genes (from a total of 5,873)(Grey bars) identified in bins (A) grouped by the Log10 of the expression level as reads per kilobase (RPK) or (B) grouped by the Log10 of raw coverage. Alternative isoforms are more likely to be detected amongst highly expressed genes compared to genes with low expression. See also additional file 9 for the relationship between read coverage and full assembly of transcripts.