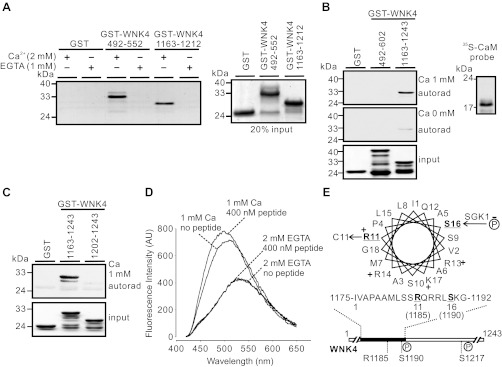
Fig. 1.
Identification of a calmodulin (CaM) binding site in the COOH-terminal region of WNK4. A: GST-WNK4 segments 492–552 and 1163–1212 exhibited ability of binding to CaM-agarose beads in the presence of 2 mM Ca2+ (left). Proteins bound to CaM-agarose beads were stained with Coomassie Blue. No binding was detected in the presence of 1 mM EGTA to chelate trace amount of Ca2+. Twenty percent of the proteins loaded in different groups were also stained by Coomassie Blue to ensure equal loading (right). B: in the presence of 1 mM Ca2+, [35S]-labeled CaM probe (autoradiograpy shown at right) bound to GST-WNK4 1163–1243, but not to GST-WNK4 492–602 (top panel). In the absence of Ca2+, both 492–602 and 1163–1243 of WNK4 lost the CaM binding ability (middle panel). Coomassie Blue staining indicated equal loading of proteins (bottom panel). C: [35S]-labeled CaM probe failed to bind to GST-WNK4 1202–1243, in which the CaM binding site was removed. Results shown in A–C are representatives from at least 3 similar experiments. D: dansyl-CaM fluorescence spectra in the absence or presence of 400 nM GST-WNK4 1163–1212 peptide at 1 mM Ca2+ or 2 mM EGTA. When dansyl-CaM interacted with the peptide in the presence of 1 mM Ca2+, the dansyl-CaM emission spectra displayed a characteristic shift in the absorption of blue light and an increase in fluorescence intensity. AU, arbitrary units. E: helical wheel projection of the CaM binding site (WNK4 1175–1192). The positively charged amino acids are indicated with “+”; disease-causing mutation site R1185 and SGK1 phosphorylation site S1190 are underlined.