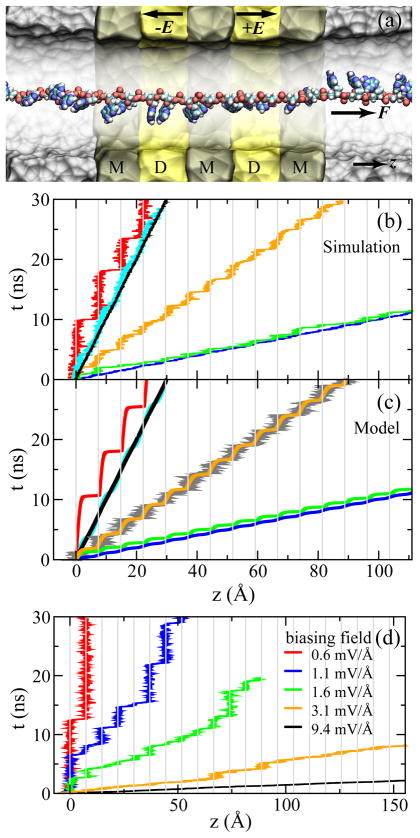
Fig. 8.
Simulations of ssDNA ratcheted through an electrostatic trap in the DNA transistor. (a) A cross-section view of the setup of MD simulations. The metal and dielectric regions are labeled with “M” and “D”, respectively. (b) Simulated time-position dependence of the ssDNA molecule pulled by a harmonic spring. (c) Time-position dependence of the ssDNA molecule that is pulled under the same conditions as in (b), resulting from the model of Eq. 2. According to the pulling velocity, data in each panel are grouped into three sets: (i) v = 1 A/ns and k = 10 (red), 100 (cyan), 1000 (black) pN/Å; (ii) v = 3 Å/ns and k = 33.3 (orange) pN/Å; (iii) v = 10 Å/ns and k = 10 (green), 100 (blue) pN/Å. (d) Thermally activated ratchet-like motion of ssDNA driven by a biasing electric field, changing from a ratcheting to a steady-sliding motion of ssDNA when increasing the biasing electric field from 0.6 mV/Å to 9.4 mV/Å, or the electric driving force on bare ssDNA from 20 to 300 pN. The interval of mesh lines in (b–d) is the interphosphate distance d. Adapted from ref. 25.