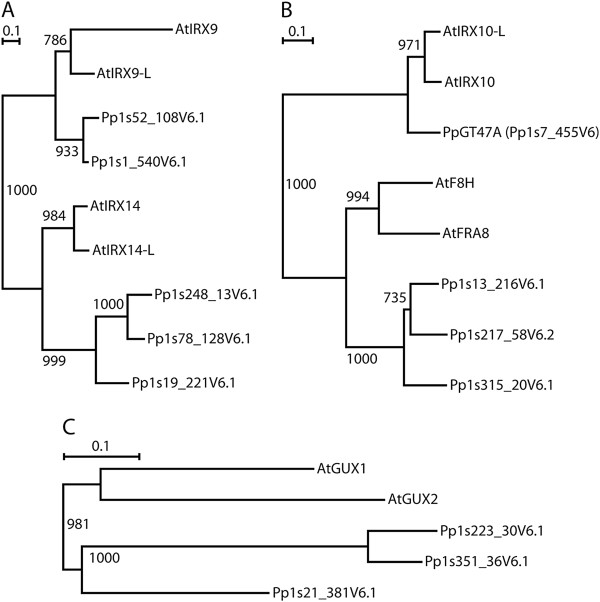
Figure 2.
Phylogenetic trees for GX related proteins in Arabidopsis and Physcomitrella. Predicted protein sequences were aligned within each family, and a tree was computed using the neighbour-joining method [46] with gapped positions being excluded and branch length corrections for multiple substitutions enabled. The bars represent a PAM value (percent accepted mutations) of 10%. The numbers shown at the branch points are bootstrap values derived from 1000 randomized sequences. A) GT43 family. B) GT47 family. C) GT8 family.