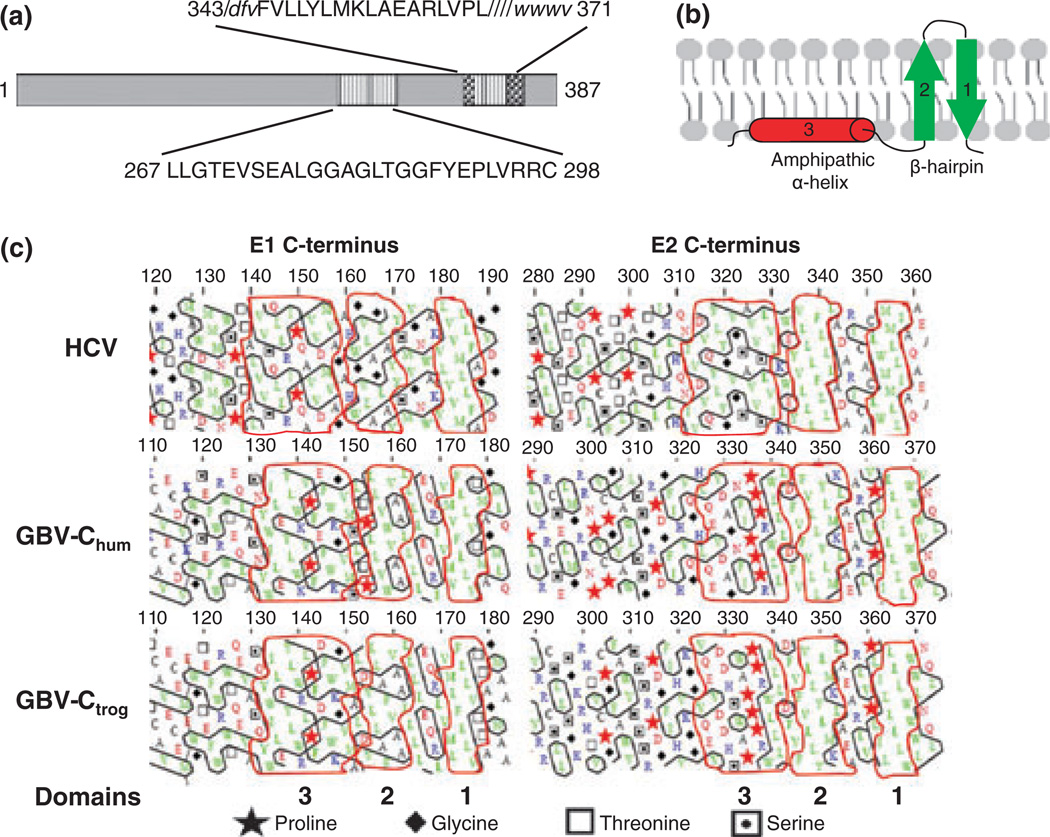
Fig. 6.
Functional mapping of the GB virus C envelope glycoprotein E2. The predicted GBV-C E2 transmembrane domain (TM) resides between amino acids 343 and 371 (a). Two putative peptide regions within E2 contain membrane interacting or fusion functions (347–364, within the TM) and 267–298. The M6 monoclonal antibody blocks E2 binding to cells, and recognizes a linear epitope including amino acids 276–292 within the putative fusion peptide (a). HCV hydrophobic TM regions on both E1 and E2 appear to be too short to serve as classical transmembrane α-helices, and are predicted to fold as two antiparallel β-strands in a hairpin structure (b). The amphiphilic α-helix upstream of the hairpin is predicted to stabilize the hairpin on the ER membrane (b). Hydrophobic cluster analysis of HCV, human and chimpanzee variants of GBV-C (GBVChum and GBV-Ctrog respectively) found that the E1 C-termini of these three viruses are similar (c) and share the putative structure shown in panel B. However, only the HCV E2 protein had this type of C terminus, while GBV-C contained helix breaking proline residues upstream from the TM region (c).