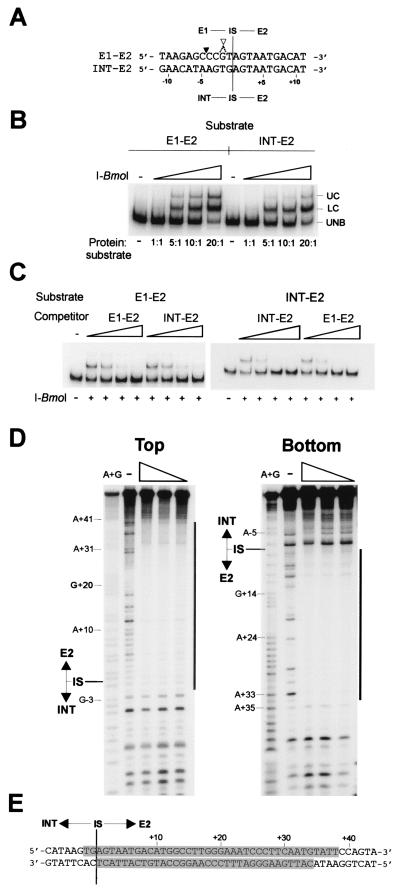
Figure 5.
I-BmoI binds with the same relative affinity to intron-containing or intronless substrates. (A) Alignment of coding strand sequence from positions −11 to +11 surrounding the intron IS of intron-containing (INT–E2) or intronless (E1–E2) substrates. I-BmoI cleavage sites are labeled as in Fig. 2. Intron-containing and intronless substrates are the same length (78 nt). (B) Gel-shift assay with increasing concentrations of I-BmoI and equal concentrations of E1–E2 or INT–E2 oligonucleotide substrate end-labeled on the top strand. (C) Intron-containing and intronless substrate can effectively compete for binding of intronless substrate by I-BmoI. Equal concentrations of intronless or intron-containing substrate end-labeled on the top strand were incubated with I-BmoI wt protein and increasing concentrations of unlabeled intron-containing or intronless substrate. (D) I-BmoI R27A protects the same region of intron-containing as intronless substrate. Shown is a DNase I protection assay with decreasing concentrations of I-BmoI R27A on top or bottom strands of INT–E2 substrate, labeled as in A. (E) Regions protected (shaded) by I-BmoI R27A from DNase I digestion on an INT–E2 substrate.