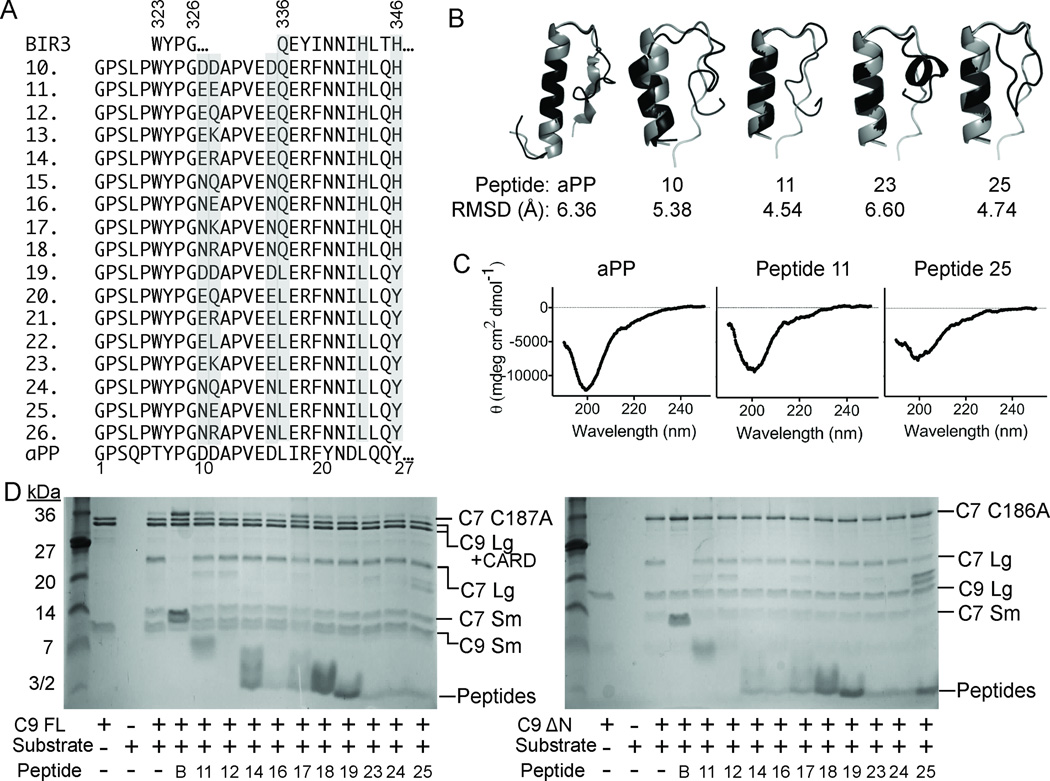
Figure 3.
Properties of aPP-scaffolded peptides. (A) Sequences of native BIR3, derivative Peptides 10–26 and native aPP, in which critical residues from the α5 region of XIAP-BIR3 have been inserted into the aPP scaffold. Highly varied positions are shaded. (B) aPP and peptide structures predicted computationally (black) by Rosetta 58, 59 are superimposed on the known structure of aPP (gray). RMSD for backbone atoms suggests the majority of the designed aPP-based peptides should adopt a helical conformation in aqueous solution. (C) The CD spectra of the aPP and Peptides 11 and 25 indicate that the structure of the designed peptides is very similar to that of native aPP in solution. (D) aPP-based peptides show some inhibition of full-length caspase-9 (C9 FL) or the N-terminal CARD-domain deleted caspase-9 (C9 ΔN) to cleave a natural caspase-9 substrate, the caspase-7 zymogen (C7 C186A) to the caspase-7 large (C7 Lg) and small (C7 Sm) subunits in an in vitro cleavage assay monitored by gel mobility.