Figure 3. Functional assays on the PHOX2B c.393_410del18 mutation (A) Reverse transcribed PCR analysis of Hela cells transfected with the respective Exontrap wild type and mutated exon 2 constructs.
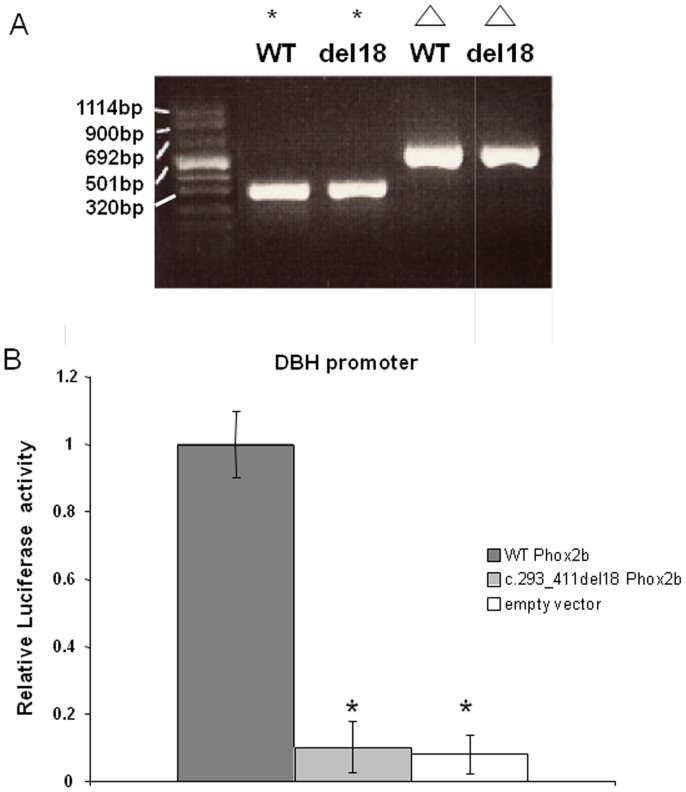
Wild type and the 393_410del18 mutated inserted human Phox2b sequences into Exontrap vectors in the correct 5′->3′ orientation have spliced their exon 2 (indicated by a triangle) as expected (approx. 500 bp, 5′ and 3′ exons of Exontrap+Phox2b exon2) demonstrating that the c.393_410del18 mutation does not seem to affect the splicing of the exon2 of Phox2b. In contrast when the exon 2 human Phox2b sequence is inverted 3′->5′ (indicated by an asterisk) within the Exontrap construct for the wild type as for the mutated sequence, the entire Phox2b insert+Exontrap-introns sequences are spliced out (approx. 300 bp). (B) Transactivation of the DBH promoter. The error bars indicate SD for n = 3 duplicate experiments. Each luciferase activity has been normalized with an internal control (pRL-CMV, Promega). In each experiment, the activity of the wild type protein was set to one, and the results are shown as the ratio between the activities of the mutant and the wild-type proteins. Significant differences between the wild-type and the specific mutant are indicated by an asterisk (Student’s test, *p<0.01).
