Figure 1. Phylogenetic tree constructed with the sequences obtained by Method A (L1 region).
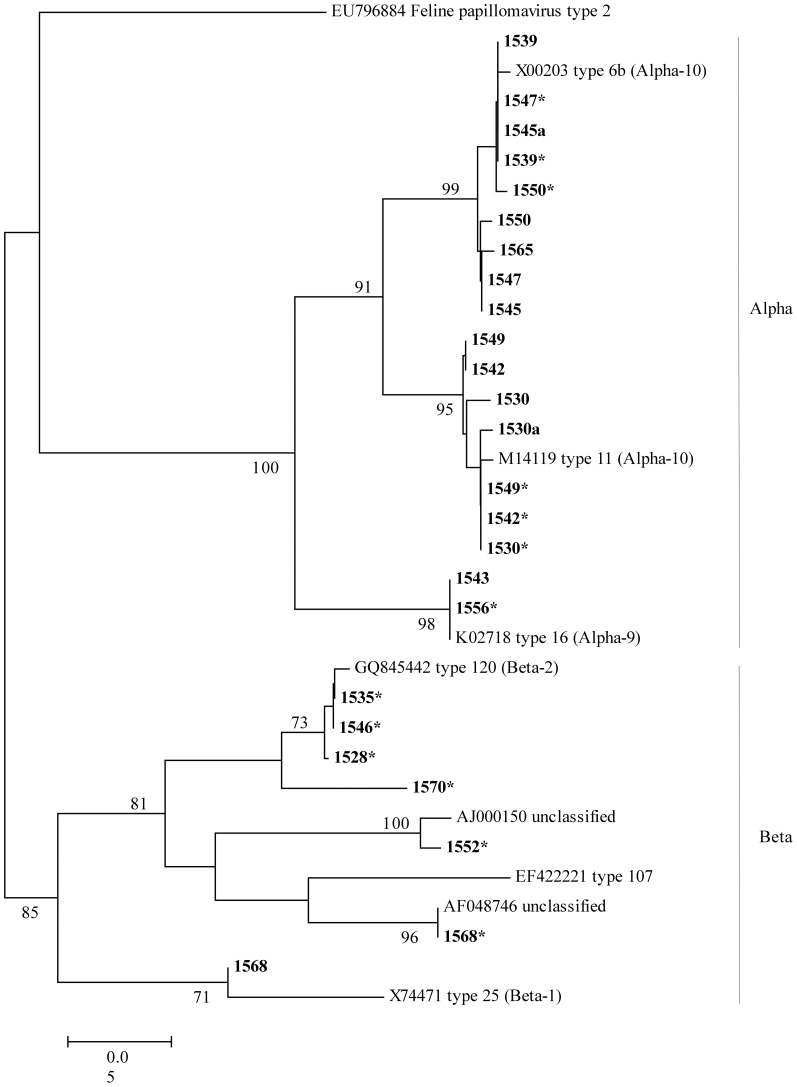
The tree was constructed using the Maximum Likelihood method based on the Kimura 2-parameter model. The sequences identified in this study are in bold. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Bootstrap values higher than 70% are shown. HPV prototypes reported in the phylogenetic tree are: HPV6b, X00203; HPV11, M14119; HPV16, K02718; HPV25, X74471; HPV120, GQ845442; AJ000150; AF048746. A feline papillomavirus type 2 (EU796884) was included as an outgroup. Identical sequences obtained from the different plasmid cloned or identical sequences obtained from the nested and hemi-nested amplicons were not included in the tree. Sequences obtained from different clones were given an alphabetical code (ID sample “a”, “b”); sequences obtained from the hemi-nested reaction were marked with an asterisk.
