Abstract
Gene targeting with genetically encoded optical voltage sensors brings the methods of voltage imaging to genetically defined neurons and offers a method of studying circuit activity in these selected populations. The present study reports the targeting of genetically encoded hybrid voltage sensors (hVOS) to neurons in transgenic mice. The hVOS family of probes employs a membrane-targeted fluorescent protein, which generates voltage-dependent fluorescence changes in the presence of dipicrylamine (DPA) as the result of a voltage-dependent optical interaction between the two molecules. We generated transgenic mice with two different high-performance hVOS probes under control of a neuron-specific thy-1 promoter. Hippocampal slices from these animals present distinct spatial patterns of expression, and electrical stimulation evoked fluorescence changes as high as 3%. Glutamate receptor and Na+ channel antagonists blocked these responses. One hVOS probe tested here harbors an axonal targeting motif (from GAP-43) and shows preferential expression in axons; this probe can thus report axonal voltage changes. Voltage imaging in transgenic mice expressing hVOS probes opens the door to the study of functional activity in genetically defined populations of neurons in intact neural circuits.
Keywords: genetic targeting, neural circuitry, voltage imaging
voltage imaging holds great promise as a technique for the study of neural circuit function. By generating real-time readouts of electrical activity from large numbers of interconnected neurons, voltage imaging can probe the emergent properties of nervous systems (Canepari and Zecevic 2010; Cohen and Salzberg 1978; Grinvald et al. 1988; Jin et al. 2002). However, conventional, synthetic, voltage-sensitive dyes stain cells with little or no specificity, forcing experimenters to contend with a superposition of signals from different neuronal and glial populations. Genetically encoded optical voltage probes offer a solution to this problem (Jin et al. 2010; Knopfel et al. 2006). They can be targeted to selected populations of cells (Huang et al. 2010; Miyoshi and Fishell 2006) to provide information about one cell type without interference from other cell types. This approach can thus overcome the lack of specificity of synthetic, voltage-sensitive dyes. Additionally, because genetically encoded voltage sensors operate through a variety of novel physical mechanisms, the genetic sensors offer an opportunity to explore strategies for improving probe performance.
The development of genetically encoded voltage sensors has followed a number of different strategies. One approach entails the incorporation of green fluorescent protein (GFP) or one of its variants into a voltage-sensitive membrane protein such as an ion channel in such a way that a structural transition in the voltage-sensing domain influences the optical properties of the fluorophore (Ataka and Pieribone 2002; Baker et al. 2007; Guerrero et al. 2002; Siegel and Isacoff 1997). Extending this approach to the incorporation of two different fluorescent proteins creates an intramolecular Förster resonance energy transfer (FRET) pair. A structural transition in the voltage-sensing domain can then generate an optical signal by altering the distance and orientation between the two fluorophores (Dimitrov et al. 2007; Sakai et al. 2001; Tsutsui et al. 2008). Although many of these probes produce signals of reasonable magnitude, a slow time response impairs their ability to detect rapid electrical events such as action potentials. Recently, a new strategy for genetically encoded voltage sensors has been introduced in which microbial rhodopsin was converted into a strongly electrochromic fluorescent protein. This Arch probe produces high-amplitude fluorescence changes but with similarly slow response times (Kralj et al. 2012). Another class of genetically encoded voltage sensors builds on an intermolecular FRET-based approach in which a synthetic fluorescent tag on the cell surface can come within range for FRET with a second fluorophore within the hydrophobic core of the lipid bilayer (Gonzalez and Tsien 1995). Because the second fluorophore is charged, voltage moves it and alters its distance to the surface tag so that the fluorescence emission of both molecules changes. This method has been converted into a genetically encoded voltage sensor by using a membrane-targeted GFP (Chanda et al. 2005). With this hybrid voltage sensor (hVOS) technique, negatively charged dipicrylamine (DPA) proved very effective as the hydrophobic charged molecule. Even though DPA is not fluorescent, its absorbance spectrum overlaps the GFP emission spectrum so that FRET can occur to quench GFP fluorescence when the two molecules are in close proximity. DPA also has the advantage of rapidly shuttling across the membrane so that hVOS has a time response of ∼0.5 ms. hVOS probes have been explored systematically and optimized to produce versions with a ∼3-fold higher sensitivity and signal-to-noise ratio than the farnesylated GFP used in the original hVOS study (Wang et al. 2010). The best of these probes detected action potentials in parts of a single cultured neuron in a single trial.
Genetically encoded voltage sensors have been tested in various types of cultured cells where expression is easy and inexpensive. However, expressing probes in intact tissue is more difficult. Furthermore, intact preparations add considerably to the background light, thus placing greater demands on probe performance. A genetically encoded voltage sensor based on voltage-sensitive phosphatase has been expressed in intact neural circuits by in utero electroporation, but experiments with this system indicated that detecting fast electrical events such as action potentials would be difficult (Akemann et al. 2010). The lack of tests for genetically encoded voltage sensors in intact neural circuits raises important questions about whether they are ready for use in the applications for which they have been designed and developed. To provide such a test, we used hVOS probes developed in this laboratory that have near optimal performance (Wang et al. 2010) to generate transgenic mice expressing these probes in neurons. These probes use cerulean fluorescent protein (CeFP) tagged at the COOH terminus with a truncated h-ras farnesylation motif. This probe will be referred to here as hVOS 1.5. Addition of a GAP-43 motif at the NH2 terminus improved performance slightly, leading to a probe referred to as hVOS 2.0. In the present study, we generated transgenic mice with both hVOS 1.5 and hVOS 2.0 under control of a neuron-specific thy-1 promoter (Feng et al. 2000). To evaluate probe performance, we used hippocampal slices, a preparation widely used in neuroscience for the study of a broad range of questions relating to learning, memory, and disease. Probe expression was quite strong in hippocampal slices, and electrical stimulation elicited robust optical responses. These results indicate that hVOS probes in transgenic mice provide a general method of imaging electrical activity from genetically defined populations of neurons in intact neural networks.
METHODS
Preparation of hippocampal slices.
Hippocampal slice preparation followed standard methods (Chang and Jackson 2006; Dean et al. 2009). Mice were decapitated under CO2-induced narcosis, as approved by the Animal Care and Use Committee of the University of Wisconsin-Madison in accordance with the guidelines of the National Institutes of Health. The brains were dissected and chilled in ice-cold cutting solution containing, in mM, 124 NaCl, 3 KCl, 26 NaHCO3, 1.25 NaH2PO4, 0.1 CaCl2, 6 MgSO4, and 10 glucose, bubbled with 95% O2-5% CO2. Transverse hippocampal slices 350 μm thick were prepared with a VT1200S Vibratome (Leica, Wetzlar, Germany). Slices were stored and studied in artificial cerebrospinal fluid (ACSF) containing, in mM, 124 NaCl, 3 KCl, 26 NaHCO3, 1.25 NaH2PO4, 2.5 CaCl2, 1.3 MgSO4, and 10 glucose bubbled with 95% O2-5% CO2. Slices were generally incubated with 4 μM DPA in ACSF for at least 1 h before hVOS experiments.
Voltage imaging.
Fluorescence was imaged with a CCD-SMQ camera (RedShirtImaging, Decatur, GA), which can acquire up to 2,000 full frames per second at a resolution of 80 × 80 pixels. An electronic shutter (VCM-D1; Uniblitz) controlled by computer limited illumination to the time of data acquisition. The camera was coupled to an Olympus BX51 fluorescence microscope with an Olympus XLUMPlanFl ×20 objective [numerical aperture (NA) = 0.95]. A stabilized 75-W Xe-arc lamp provided illumination (Opti Quip, Highland Mills, NY). An Olympus filter cube for enhanced cyan fluorescent protein (ECFP) was used for hVOS; an Olympus rhodamine filter cube was used to image the voltage-sensitive, fluorescent dye di-4-ANEPPDHQ (Invitrogen, Grand Island, NY), but the emission filter was replaced with a 600- to 700-nm band-pass filter (Chroma Technology, Brattleboro, VT).
Stimulating electrodes with 10- to 30-μm tip diameters were fabricated from borosilicate glass capillaries (1.15-mm inside diameter, 1.50-mm outside diameter) and filled with ACSF. Slices were stimulated with 0.18-ms, 200-μA current pulses delivered by a monopolar constant-current stimulus isolator (Model A365; World Precision Instruments, Sarasota, FL). Data acquisition, stimulation, and shutter were all controlled by Neuroplex software provided with the CCD-SMQ camera.
For imaging with di-4-ANEPPDHQ, slices were stained for 20 min in ACSF containing 7 μg/ml dye.
Transgenic mice.
We used two previously described hVOS probes (Wang et al. 2010), hVOS 1.5 (CeFP with a truncated h-ras motif at the COOH terminus) and hVOS 2.0 (CeFP with the same C-terminal tag and with a motif from GAP-43 at the NH2 terminus). DNA encoding these two probes was cloned into a 6.5-kb expression cassette derived from the mouse thy-1.2 gene containing the promoter to the intron and exons 1, 2, and 4 (Feng et al. 2000). This construct lacks exon 3 and its translation initiation site, and this deletion eliminates expression in nonneuronal cells. hVOS probe encoding sequences were inserted at the XhoI site by blunt-end ligation, digested with EcoRI and Pvu I, and purified. Transgenic mice were then generated by the Transgenic Animal Facility in the Biotechnology Center of the University of Wisconsin-Madison (http://www.biotech.wisc.edu/facilities/transgenicanimal). DNA was injected into one-cell FVB mouse embryos, and the embryos were surgically implanted into pseudopregnant females. Offspring were genotyped for the transgene at weaning, and genotype-positive founders were bred with wild-type mice to generate transgenic lines.
Two-photon microscopy.
Two-photon fluorescent micrographs were taken with an Ultima scanning system (Prairie Technologies, Middleton, WI) coupled to an Olympus BX61 microscope with an Olympus XLUMPlanFl N ×20 objective (NA = 1.0). A Coherent Chameleon Ti:Sapphire laser provided illumination. In Fig. 1, multiple images were merged using the automerge feature of the computer program Photoshop.
Fig. 1.
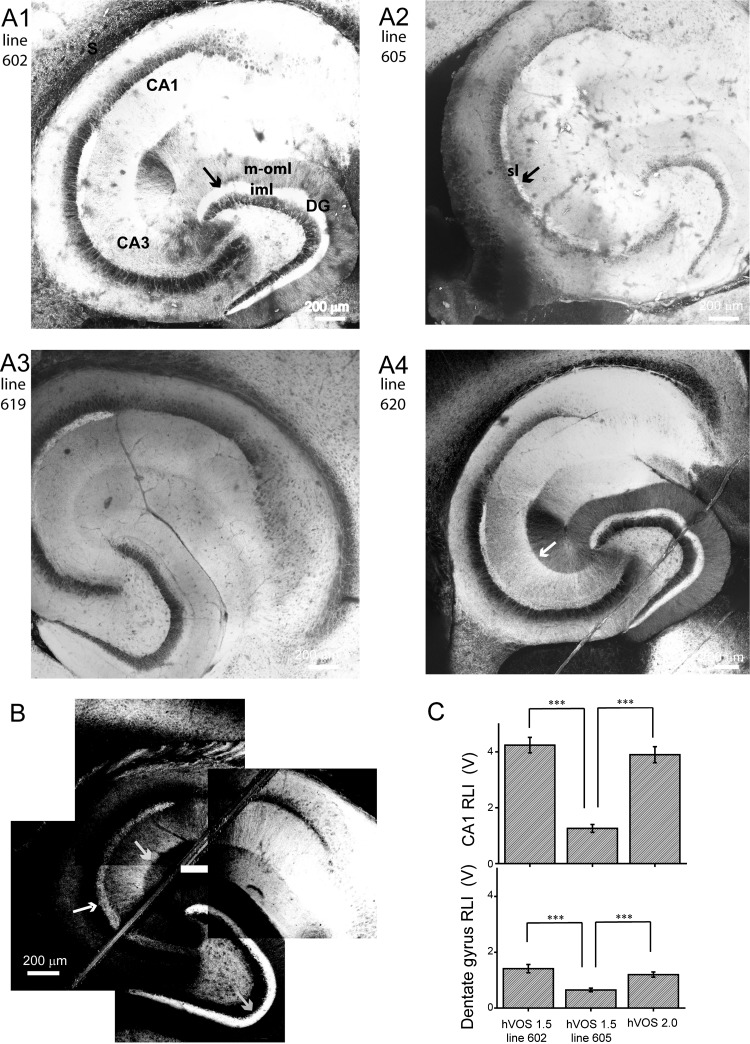
Broad patterns of probe expression in hippocampus. Hippocampal slices were prepared from neuron-specific promoter thy-1-hybrid voltage sensor 1.5 (hVOS 1.5; A) and thy-1-hVOS 2.0 (B) mice and viewed with a 2-photon microscope. Montages were constructed from images of multiple fields of view to illustrate overall patterns across entire slices. A1: hVOS 1.5 line 602 shows high probe expression in the inner molecular layer (iml) of the dentate gyrus (DG; arrow) and relatively uniform expression within the dendritic layers. Labels indicate CA1 and CA3 regions, subiculum (S), and middle-outer molecular layer (m-oml). A2: line 605 shows diffuse expression throughout the slice, with higher expression in the stratum lucidum (sl; arrow). A3: line 619 has a similar expression pattern as line 605 (B). A4: line 620 has a similar expression pattern as line 602 (A1). In the CA3 region, a sharp border separates the brighter stratum radiatum (sr) and dimmer stratum lacunosum-moleculare (sl-m; white arrow). This feature is present in line 602 but is less prominent. B: slices from the 1 hVOS 2.0 line showed especially strong expression in the sl (white arrow) and iml (gray arrow, bottom right). As in hVOS 1.5 lines 602 and 620, a sharp border can be seen between the sr and sl-m (gray arrow, middle). A1–A3 show montages of 4 × 4 images; A4 shows a montage of 5 × 5 images; these panels were merged with the automerge feature of Photoshop. B shows a montage of 4 fields that left a small empty white rectangle in the center. A nylon harp was used to hold slices down, and its fibers are visible in A4 and B. C: resting light intensities (RLI) from the CA1 region and DG averaged over 10–14 slices. RLI was averaged over 250- × 250-μm regions in each slice. All measurements were made with the same illumination intensity. ***P < 0.001 by Student's t-test.
Reagents.
All chemicals and drugs were purchased from Sigma-Aldrich.
Software and analysis.
hVOS imaging data were acquired with a computer running Neuroplex, the program provided by RedShirtImaging with the CCD-SMQ camera. With this program, we specified the stimulation protocol, data acquisition rate, spatial resolution, and shutter control. In the present study, CCD-SMQ images were displayed as either 10- × 10- or 80- × 80-pixel images. Neuroplex also performed baseline corrections and high- and low-pass filtering. Traces from individual locations on 10- × 10-pixel grids were exported to the computer program Origin for display and analysis.
Means are presented ± SE, and differences were evaluated for statistical significance using Student's t-test.
RESULTS
For hVOS 1.5, we identified four transgenic-positive lines that expressed probe in brain and bred well: lines 602, 605, 619, and 620. Three hVOS 2.0 lines were transgenic-positive, but only one bred well and expressed the fluorescent protein strongly in brain. We report the results for the four hVOS 1.5 mice and one hVOS 2.0 mouse as they are viable, breed normally, and show no obvious differences in behavior from wild-type mice. They all express probe in hippocampus, and we first summarize the expression patterns before presenting results on the imaging of electrical activity.
Probe expression patterns.
Two-photon microscope images of hippocampal slices from each of the four hVOS 1.5 transgenic lines reveal probe expression with distinct spatial patterns (Fig. 1, A1–A4). hVOS 2.0 expression also showed a highly distinctive spatial pattern (Fig. 1B). In general, these expression patterns were rich in detail, reflecting differences in neuronal cell types as well as differences in the morphology and process architecture of those cell types. We note the following features in probe expression.
Low-magnification images of large areas provided an overall view of expression throughout a hippocampal slice. Fluorescence in the CA1, CA2, and CA3 regions indicate that pyramidal cells in all five lines expressed probe. Slices from hVOS 1.5 lines 602 (Fig. 1A1) and 620 (Fig. 1A4) had a number of features in common. Fluorescence was strong in the dentate gyrus (DG), especially in the inner molecular layer (iml), with a sharp boundary between the iml and middle-outer molecular layer (m-oml). Slices from the other two hVOS 1.5 lines, 605 (Fig. 1A2) and 619 (Fig. 1A3), had similar expression patterns to one another, and these patterns differed from those seen in lines 602 and 620. Notably, their fluorescence was nearly uniform in the molecular layer with only a very faint boundary between the iml and m-oml. Slices from all four hVOS 1.5 lines had fluorescence in the stratum lucidum (sl), which was brighter than the surrounding regions. In the CA3 region, slices from hVOS l.5 line 620 displayed a distinct boundary between the stratum radiatum (sr) and stratum lacunosum-moleculare (sl-m; Fig. 1A4, white arrow); a fainter boundary was seen in line 602 (Fig. 1A1). This pattern suggests probe expression in Schaffer collaterals arising from CA3 pyramidal cells. Fluorescence is fainter in the sl-m due to the absence of these axons, but this region also contains CA3 pyramidal cell dendrites, and the residual fluorescence we see arises from these structures. Transgenic lines 602 and 620 displayed especially bright fluorescence in the subiculum. This likely reflects the dense arborization of CA1 pyramidal cell axons in this region (Amaral et al. 1991), although expression in the pyramidal cells of the subiculum is also possible.
The bright fluorescence in the iml with hVOS 1.5 lines 602 and 620 (Fig. 1, A1 and A4) suggests that these lines have strong probe expression in hilar mossy cells, as these cells form synapses on dentate granule cells in a dense projection tightly confined to the iml (Amaral et al. 2007). In hVOS 1.5 lines 605 and 619 (Fig. 1, A2 and A3), the entire molecular layer showed nearly uniform expression. The absence of a distinct bright band in the iml indicates that in this transgenic line the probe does not target mossy cells but is distributed uniformly in granule cell dendrites. Furthermore, the expression pattern is consistent with the absence of probe in perforant-path axons, which terminate in the m-oml and would make these layers brighter than the iml.
The spatial pattern of probe expression in slices from our one hVOS 2.0 line (Fig. 1B) broadly resembled that seen in slices from hVOS 1.5 lines 602 and 620 but had stronger expression in the sl and iml. However, the m-oml, stratum oriens (so), sr, as well as the cell body layers, stratum pyramidale (sp) and stratum granulosum (sg), displayed much lower probe levels than all of the hVOS 1.5 lines. These features reflect a trend of preferential hVOS 2.0 expression in axons. This point will be discussed further below.
To gauge fluorescence quantitatively, we compared the resting light intensity in 10 or more slices of each transgenic line under identical conditions using the CCD-SMQ camera used in imaging experiments. Figure 1C presents these results from 250- × 250-μm fields in the CA1 region and DG of slices from hVOS 1.5 lines 602 and 605 and hVOS line 2.0 (Fig. 1C). These show general trends of >2-fold higher probe expression in the CA1 region than the DG and comparable expression between hVOS 1.5 line 602 and the hVOS 2.0 line. Expression in both of these lines was greater than in hVOS 1.5 line 605.
Higher-magnification two-photon microscope images provided a more detailed comparison of expression patterns (Fig. 2). Dendritic localization is evident in the m-oml in hVOS 1.5 line 602 (Fig. 2A1) but much weaker in hVOS 2.0 (Fig. 2C1). Thus the hVOS 1.5 probe is expressed more strongly in granule cells dendrites compared with hVOS 2.0. hVOS 2.0 shows less expression in granule cell dendrites, but granules cells clearly express hVOS 2.0 as judged by the striking fluorescence in the sl (Figs. 1B and 2C2), which contains mossy fibers originating from granule cell axons. Therefore, the comparison of expression in the sl with expression in the molecular layer in these two lines indicates that hVOS 2.0 preferentially targets axons. Additional indications of preferential axonal targeting by hVOS 2.0 can be found in the CA3 region, where the sp shows some expression in the hVOS 1.5 lines (Fig. 2, A2 and B2) but is much darker in the hVOS 2.0 line (Fig. 2C2). Nerve terminal expression of hVOS 2.0 also appears in the subiculum (Fig. 2C3). The axons in slices from hVOS 1.5 lines do not show such clear tubulelike or boutonlike structures as slices from the hVOS 2.0 line. These distinctions in expression illustrate a broad trend of preferential hVOS 2.0 expression in axons. The dendrites and cell bodies generally show stronger fluorescence in slices from hVOS 1.5 animals than hVOS 2.0. The preferential expression of hVOS 2.0 in axons presumably reflects our incorporation into this probe of an axonal targeting motif from GAP-43 (El-Husseini et al. 2001; Mosevitsky 2005). Lacking this motif, hVOS 1.5 distributes more evenly in cell bodies, dendrites, and axons.
Fig. 2.
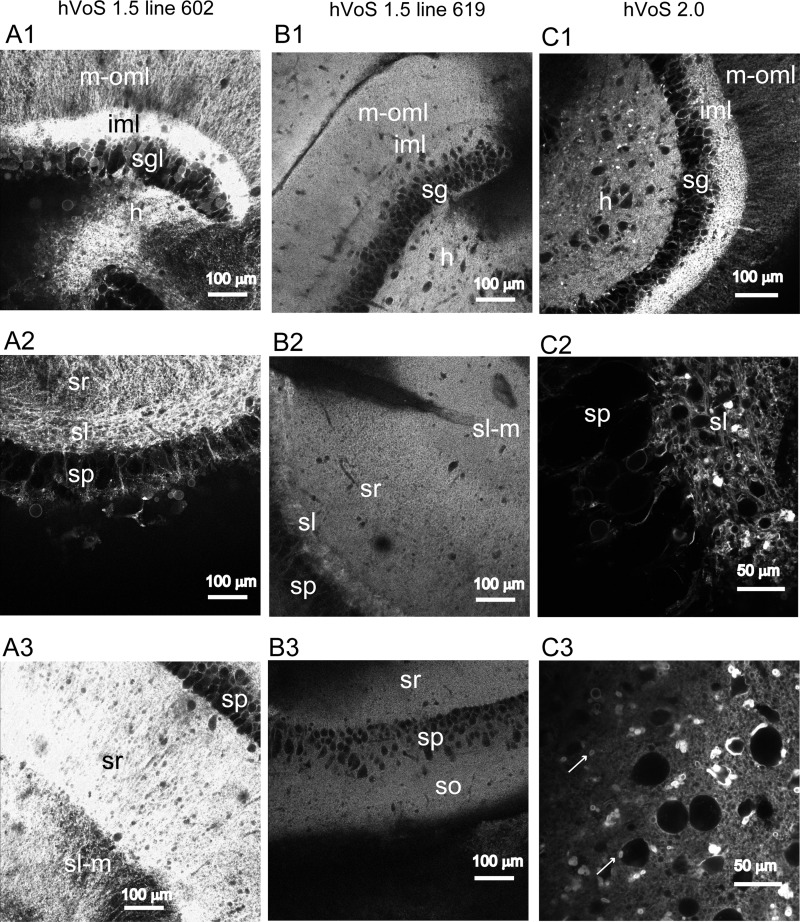
Cellular localization of hVOS probes. Two-photon microscope images at higher magnification than in Fig. 1 show the locations of hVOS probes in slices from hVOS 1.5 line 602 (A1–A3), hVOS 1.5 line 619 (B1–B3), and the hVOS 2.0 line (C1–C3). Row 1 compares the DG of these lines; row 2 compares the CA3 region; row 3 compares the CA1 region between the 2 hVOS 1.5 lines (A and B) but for the hVOS 2.0 line (C3) displays the subiculum. In the DG, different levels of probe expression are evident in the hilus (h), stratum granulosum (sg, sgl), iml, and m-oml. In the CA3 region, different levels of probe expression are evident in the stratum pyramidale (sp), sr, sl-m, and sl. Note that the stratum oriens (so), which strongly expresses probe in line 602 (Fig. 1A1), is out of the plane of focus in this image. In the CA1 region, probe expression is evident in the so, sp, sr, and sl-m (A3 and B3). Comparing A1 and C1 shows that both transgenic lines have high probe expression in iml and lower but significant expression in h. hVOS 2.0 expression is punctate in the sl (C2) and subiculum (C3; arrows), indicative of nerve terminal localization.
Slices from all four hVOS 1.5 lines displayed fluorescence both in the m-oml, which contains granule cell dendrites, and the sl, which contains granule cell axons. Thus expression in granule cells appears to be a general feature of these lines. In addition, two lines, 602 and 620, present probe in the iml, indicating expression in hilar mossy cells. Their pattern can thus be viewed as a superposition of expression in both granule cells and mossy cells. The hVOS 2.0 pattern is a variation on this dual granule cell/mossy cell motif but with higher expression in axons relative to dendrites and cell bodies.
hVOS signals.
In hippocampal slices from all four hVOS 1.5 lines and the hVOS 2.0 line, electrical stimulation evoked clear fluorescence changes provided that DPA was present (Fig. 3). We generally used 4 μM DPA as this concentration provides excellent signals and produces minimal pharmacological action (Wang et al. 2010). A CCD-SMQ image of a slice from hVOS 1.5 line 602 with fluorescence traces overlain (Fig. 3A) shows no detectable changes in fluorescence on electrical stimulation in DPA-free ACSF. Two hours after changing to ACSF with 4 μM DPA, the same stimulation evoked responses throughout the sr of the CA1 region. The fluorescence changes developed in ∼2 ms and then decayed to baseline in ∼50 ms (Fig. 3A, right, shows selected traces). The time courses of these signals qualitatively resemble those seen with conventional voltage-sensitive dyes in hippocampal slices (Chang and Jackson 2006; Grinvald et al. 1982; Jackson and Scharfman 1996), and this comparison will be examined quantitatively below.
Fig. 3.
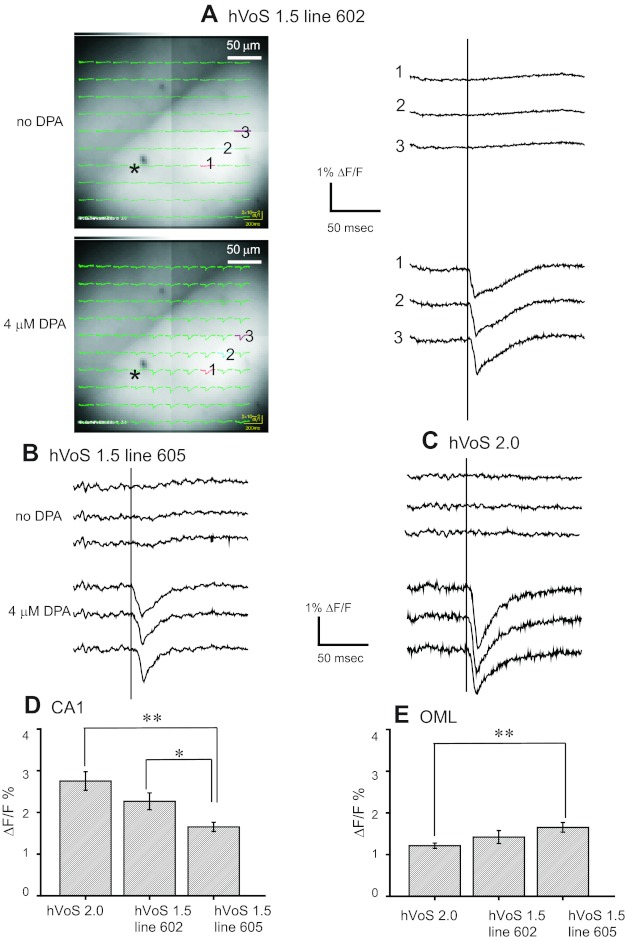
Stimulus-evoked fluorescence changes (ΔF/F) and their dipicrylamine (DPA) dependence. Slices from 2 hVOS 1.5 lines and the hVOS 2.0 line were tested 1st in control solutions and then 2 h after addition of 4 μM DPA. All recordings were from the sr of the CA1 region stimulated with 200-μA, 0.18-ms pulses. A, left: a slice from hVOS 1.5 line 602 with images and fluorescence traces superimposed. Right: traces from the 3 numbered locations before (top 3) and 2 h after (bottom 3) addition of DPA. No hVOS signals were seen in the absence of DPA; after addition of 4 μM DPA, responses were seen throughout the sr. The site of stimulation is indicated by the asterisk, and the out-of-focus stimulation electrode is faintly visible to the left. B: traces from an experiment similar to A (i.e., in the CA1 region with the same stimulation parameters) are displayed for hVOS 1.5 line 605 before and after addition of DPA. C: traces from an hVOS 2.0 slice before and after addition of DPA (similar to A and B). Bar graphs display mean stimulus-evoked fluorescence changes in 4 μM DPA in each of the 3 indicated transgenic lines in the CA1 region (D) and the oml of the DG (E). All responses were evoked by 200-μA, 0.18-ms pulses. Error bars are SE with 9–12 measurements each. *P < 0.05; **P < 0.01 by Student's t-test. All traces were averages of 10 trials.
hVOS probe fluorescence decreases with membrane depolarization because DPA moves to the inner surface of the cell membrane where hVOS probes are anchored; the arrival of DPA quenches the probe fluorescence. Responses of approximately 1–3% could be seen throughout the field of view, and amplitudes are summarized in Fig. 3, D and E. We averaged 10 trials from single sites in 10- × 10-pixel images (25 × 25 μm) to compare transgenic lines and regions using data collected under identical conditions. Responses were generally visible in single trials, and Fig. 7, D and E, shows examples of very clear single-trial responses. Traces from the CA1 region of hVOS 1.5 line 605 (Fig. 3B) and the hVOS 2.0 line (Fig. 3C) were similar. Although increasing the stimulation pulse duration to 2 ms often evoked fluorescence changes near the site of stimulation in the absence of DPA, we only saw a small DPA-independent fluorescence change in one instance with the stimulus parameters used in the present study, and this signal was restricted to the site of stimulation.
Fig. 7.
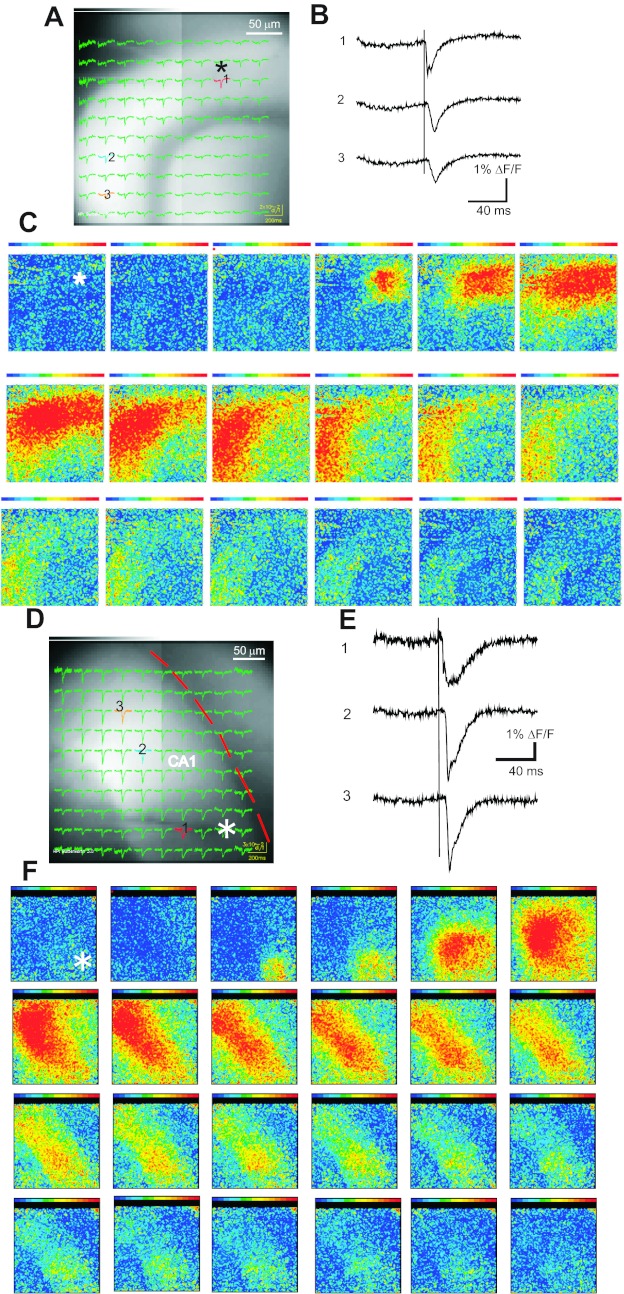
Signal spread in the DG and CA1 region. A: the DG of a slice from hVOS 1.5 line 605 with hVOS traces superimposed. The slice was stimulated in the molecular layer (asterisk). B: traces from locations indicated in A (10 trials were averaged). C: a sequence of maps with fluorescence encoded as color (red strongest, purple weakest) at 2-ms intervals shows the spread of signals through the molecular layer (site of stimulation indicated with asterisk). D: the CA1 region is shown in a slice from the hVOS 2.0 line with fluorescence traces superimposed. The sp is highlighted by the red curve, and the stimulation electrode is faintly visible (asterisk). E: selected traces from locations indicated in D (single-trial responses without averaging). F: a sequence of maps with intensity encoded as color show the pattern of signal propagation at 2-ms intervals (site of stimulation indicated with asterisk). Stimulation, 0.18 ms, 200 μA; [DPA] = 4 μM.
Stimulus-evoked hVOS signals varied with location as well as with transgenic line. Signal amplitudes for two locations and three different transgenic lines are summarized in Fig. 3, D and E, for the same stimulus protocol. The largest responses were seen in the CA1 region in slices from the hVOS 2.0 line where the average fluorescence change was ΔF/F = 2.8% (Fig. 3D). These signals were larger than those seen with hVOS 1.5 (ΔF/F = 2.2% in line 602 and 1.6% in line 605). However, in the oml of the DG, the hVOS 2.0 signals of 1.1% were smaller than the hVOS 1.5 signals of 1.4% in line 602 and 1.7% in line 605 (Fig. 3E).
The largest hVOS 2.0 signals of ∼3% were ∼1/3 as large as the signals seen in cultured neurons expressing this probe during an action potential with 2 μM DPA (Wang et al. 2010). The magnitude of the voltage change in the present experiments is unknown. Pharmacological experiments presented below indicate that the signals reflect a composite of action potentials and synaptic potentials (Figs. 4 and 5). Fluorescence from neurons not activated by the stimulus as well as imperfect synchrony of the population responses will reduce the peak fluorescence change in the present experiments. Slice autofluorescence will add to the background and further reduce the magnitude of a signal in slices. Thus the amplitudes seen in a slice measurement should be smaller than those seen in cultured neurons, and the ∼3-fold reduction reflects a combination of these various factors.
Fig. 4.
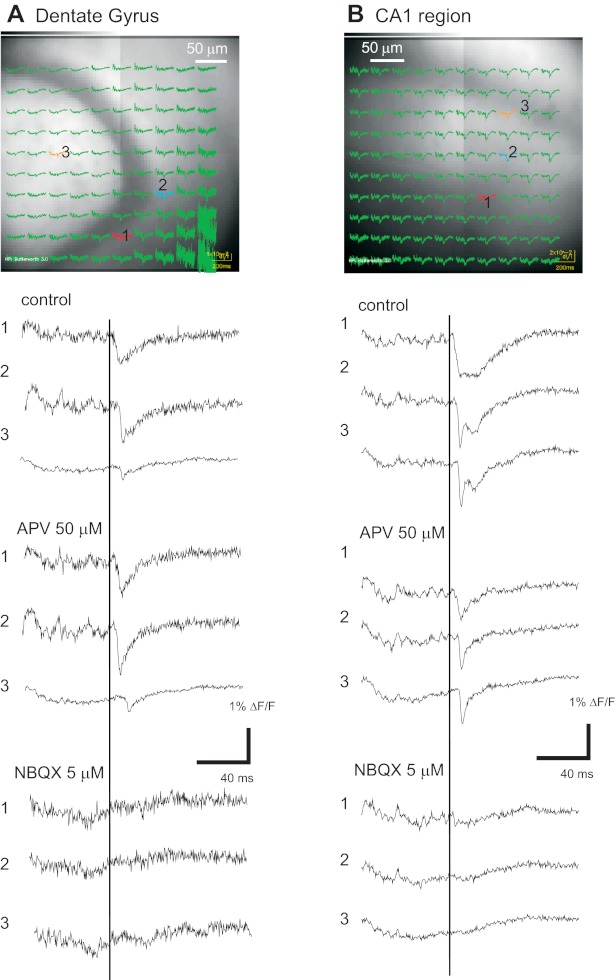
Glutamate receptor dependence of hVOS 1.5 signals. hVOS signals in the DG and CA1 region in hippocampal slices from hVOS 1.5 line 605. A: image of the DG with hVOS traces superimposed for a control recording before drug addition. Three selected traces from locations indicated by numbers show control responses and responses after addition 1st of 50 μM amino-5-phosphonovaleric acid (APV) and then 5 μM 6-nitro-2,3-dioxobenzoquinoxaline sulfonamide (NBQX; with continued presence of APV). B: images from the CA1 region with hVOS traces superimposed. Selected traces below the image show the same sequence of drug addition as A. The responses in APV decayed faster than in control and were completely blocked by NBQX. All responses were evoked by 200-μA, 0.18-ms pulses applied to location 1 (sg in the DG and the sr in the CA1 region). [DPA] = 4 μM; all traces were averages of 10 trials.
Fig. 5.
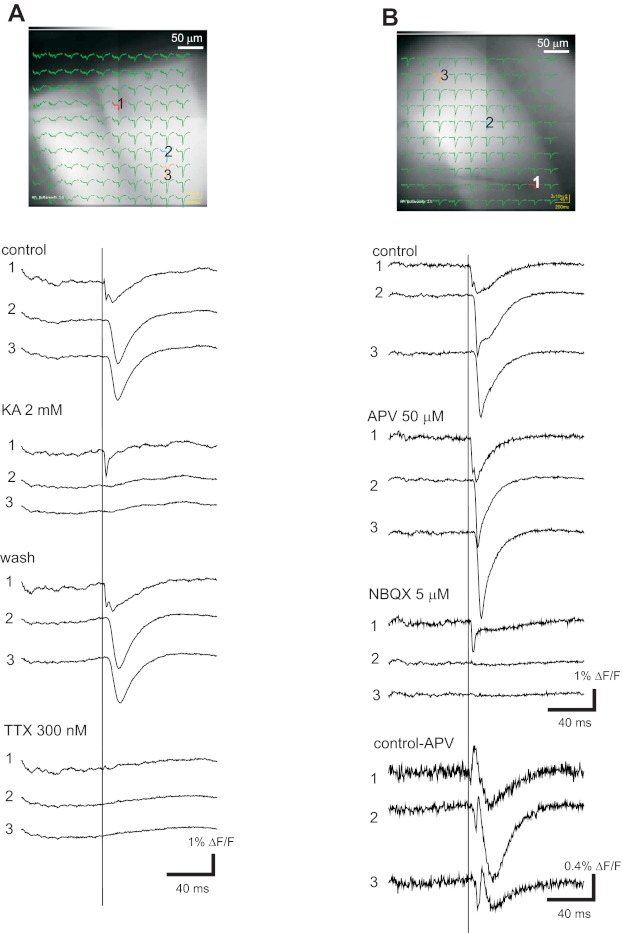
Glutamate receptor and Na+ channel dependence of hVOS 2.0 signals. Images of the CA1 region with traces superimposed are shown for 2 experiments, with traces from the indicated locations below the images. A: control traces show responses before drug addition; 2 mM kynurenic acid (KA) blocked responses except in traces close to the stimulation site. Washing out drug reversed the block. TTX (300 nM) blocked responses including those close to the stimulation site. B: control traces show responses before drug addition; 50 μM APV blocked N-methyl-d-aspartate (NMDA) receptors, revealing the dl-α-amino-3-hydroxy-5-methylisoxazole-propionic acid (AMPA) receptor-dependent component; subsequent addition of 5 μM NBQX (with continued presence of APV) blocked the AMPA receptor-mediated responses, leaving only a local spike near the site of stimulation. Scale bars near NBQX traces apply to control and APV. The bottom 3 traces are differences formed by subtraction of the traces in APV from control traces to show the NMDA receptor-mediated component. These traces are displayed with an expanded scale. All responses were evoked by 0.18-ms, 200-μA pulses at location 1. [DPA] = 4 μM; all traces were averages of 10 trials.
It is notable that hVOS 2.0 signals are much smaller in the DG. hVOS 1.5 signals from two different lines also produced weaker signals in the DG, although the difference was much smaller, and in the case of line 605 the difference was negligible. hVOS 1.5 signals for line 605 were significantly smaller than hVOS 2.0 signals in the CA1 region (Fig. 3D) and significantly larger in the DG (Fig. 3E). Some of these variations in relative signal strength are likely to reflect the different subcellular locations of the probe. In line 602 and 605, hVOS 1.5 is clearly expressed in the granule cell dendrites (Fig. 1, A1 and A2). By contrast, the strong hVOS 2.0 fluorescence in the DG is confined to the iml where mossy cell axons are located. The better performance of hVOS 1.5 in the DG can thus be explained by the greater activation of granule cell dendrites than of mossy cell axons and the lower levels of hVOS 2.0 in dendrites.
Pharmacological sensitivity of hVOS signals.
Experiments with receptor antagonists established that responses evoked by electrical stimulation depend on the activation of glutamate receptors for both hVOS 1.5 (Fig. 4) and hVOS 2.0 (Fig. 5). In slices from hVOS 1.5 line 605, signals in the DG were reduced slightly by the N-methyl-d-aspartate (NMDA) receptor antagonist amino-5-phosphonovaleric acid (APV; 50 μM). After APV addition, responses decayed more rapidly (Fig. 4A), as expected from the more rapid kinetics of dl-α-amino-3-hydroxy-5-methylisoxazole-propionic acid (AMPA) receptors. The remaining response was essentially completely blocked by the AMPA receptor antagonist 6-nitro-2,3-dioxobenzoquinoxaline sulfonamide (NBQX; 5 μM; Fig. 4A). In the CA1 region, APV blocked a larger fraction of the evoked response, indicating that NMDA receptors make a greater contribution to responses in this region (Fig. 4B). Again, NBQX completely blocked the residual response seen in the presence of APV.
In the CA1 region of slices from the hVOS 2.0 line, the broad-spectrum glutamate receptor antagonist kynurenic acid (KA; 2 mM) blocked responses completely except in the immediate vicinity of the stimulation electrode (Fig. 5A). Washout of KA allowed responses to recover (Fig. 5A, wash). After recovery, application of TTX blocked responses completely, including responses in the vicinity of the stimulation electrode (Fig. 5A, TTX). This indicates that near the stimulation electrode, responses seen in the presence of KA depend on voltage-gated Na+ channels. The sequence of APV followed by NBQX had the same effect in the CA1 region of slices from the hVOS 2.0 line (Fig. 5B) as in slices from hVOS 1.5 line 605 (Fig. 4B); responses in APV decayed more rapidly, and NBQX blocked the residual response. Subtraction of responses in APV from predrug control responses revealed the response component mediated by NMDA receptors, which has a slow time course characteristic of the kinetics of these receptors (Fig. 5B, bottom traces). These experiments indicate that signals detected with both hVOS 1.5 and 2.0 reflect excitatory synaptic activity and action potentials.
Comparison with di-4-ANEPPDHQ.
To compare hVOS signals with another voltage-imaging probe, we tested three widely employed, synthetic, voltage-sensitive dyes, RH414, di-4-ANEPP, and di-4-ANEPPDHQ (Obaid et al. 2004), in the presence of DPA. Di-4-ANEPPDHQ produced the largest signals with the highest signal-to-noise ratio, but DPA inverted the sign of the stimulus-induced changes in di-4-ANEPPDHQ fluorescence. Figure 6 displays hVOS and di-4-ANEPPDHQ signals recorded alternately from the same sites in the CA1 region by switching filter sets. The downward fluorescence changes with hVOS imaging are accompanied by upward fluorescence changes with di-4-ANEPPDHQ. In the absence of DPA, stimulus-evoked di-4-ANEPPDHQ fluorescence changes were upward (data not shown). This reversal of these signals following the addition of DPA is likely the result of a FRET interaction between DPA and di-4-ANEPPDHQ, similar to that described between DPA and the voltage-insensitive fluorescent dye DiO (Bradley et al. 2009).
Fig. 6.
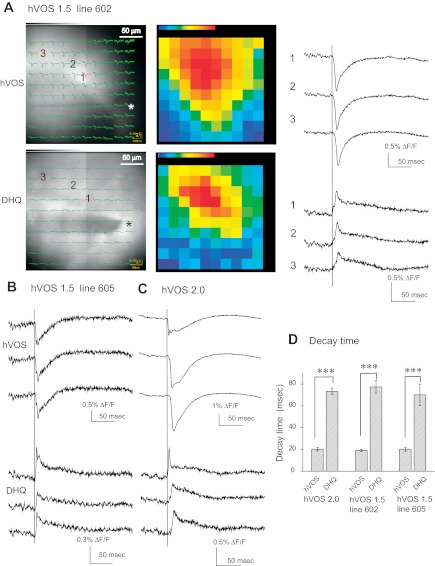
hVOS and di-4-ANEPPDHQ (DHQ). A: images and superimposed traces from an hVOS 1.5 line 602 slice (site of stimulation indicated with asterisk). The top shows hVOS signals superimposed on the charge-coupled device (CCD) image (right) and maps with the maximal signal at each location encoded as color (red strongest, purple weakest). The bottom shows the corresponding results for the synthetic, voltage-sensitive, fluorescent dye di-4-ANEPPDHQ (stained with 7 μg/ml dye in ACSF for 20 min). The distributions of fluorescence and of evoked fluorescence changes are different for hVOS and di-4-ANEPPDHQ, but the data were taken from the same field of view with the same stimulation by switching filter sets. Traces to the right of the images show stimulus-evoked fluorescence changes for hVOS (top) and di-4-ANEPPDHQ (bottom). Note that in the absence of DPA, di-4-ANEPPDHQ signals are downward (Obaid et al. 2004), and this was confirmed here (data not shown); DPA has a Förster resonance energy transfer (FRET) interaction with di-4-ANEPPDHQ that inverts the signals. B: hVOS and di-4-ANEPPDHQ traces from the same locations in a slice from an hVOS 1.5 line 605 mouse. C: hVOS and di-4-ANEPPDHQ traces from a slice from an hVOS 2.0 mouse. D: response decay times for hVOS and di-4-ANEPPDHQ for 10 or more experiments with each of the transgenic lines displayed in A–C. Decay time was taken as the time to fall to 37% of the peak. All responses were from the same part of the CA1 region, with 0.18-ms, 200-μA stimulation pulses. [DPA] = 4 μM; all traces in A–C are averages of 10 trials. ***P < 0.001 by Student's t-test.
In most cases, the hVOS signal amplitude and signal-to-noise ratio were clearly better than for di-4-ANEPPDHQ (Fig. 6, A and C). The one case where the advantage of hVOS was less clear was with hVOS 1.5 line 605 (Fig. 6B), and this probe had the weakest hVOS signal in the CA1 region (Fig. 3D). With both hVOS 1.5 and di-4-ANEPPDHQ, optical responses to electrical stimulation spread over considerable distances, as revealed by the traces superimposed on images (left column of Fig. 6A) and the color maps of maximum response (middle column of Fig. 6A). The spread was in the same direction with both probes; the site of stimulation was in the bottom right corner of the imaged region in Fig. 6A, and the color maps show upward spread. The area of the activated region appeared to be greater with hVOS 1.5.
Very similar results were seen with di-4-ANEPPDHQ in hVOS 1.5 lines 602 and 605 as well as the hVOS 2.0 line (Fig. 6, B and C). Response latency showed a parallel increase with distance for both the hVOS and synthetic probes. All hVOS and di-4-ANEPPDHQ responses rose in ∼2 ms, but the di-4-ANEPPDHQ signal decayed more slowly than the hVOS signals in all three hVOS lines tested (Fig. 6, A–C). For all three lines, the time to decay by 37% was ∼4-fold shorter for hVOS than di-4-ANEPPDHQ (Fig. 6D). The slower decay with the synthetic dye is similar to that noted for another synthetic dye, RH155, which reports voltage from both neurons and glia (Kojima et al. 1999; Konnerth et al. 1987). The slower voltage changes arising from glial glutamate transporter currents can thus account for the difference between the decay times of hVOS and di-4-ANEPPDHQ. This illustrates a benefit of genetic targeting in focusing on signals arising from neurons. The presence of di-4-ANEPPDHQ in glia may also be relevant to the less extensive spread of responses that we see with this probe (color maps in Fig. 6A).
hVOS responses and hippocampal circuitry.
Slices from hVOS 1.5 line 605 exhibited strong and relatively uniform fluorescence in the molecular layer of the DG (Fig. 1A2). Stimulation in the molecular layer of a slice from this line evoked responses throughout the molecular layer, as indicated by traces overlain on the slice image (Fig. 7A). Traces from locations further from the site of stimulation show longer latencies reflecting propagation through the DG circuitry (Fig. 7B). The propagation is depicted with a sequence of color maps at 2-ms intervals with signal amplitude encoded as color (Fig. 7C). The responses were initiated at the site of stimulation (site 1 in Fig. 7A) and spread throughout the molecular layer within 10–15 ms. This result recapitulates behavior observed in the DG in rat hippocampal slices stained with a synthetic dye (Jackson and Scharfman 1996; Scharfman et al. 2002). These studies showed that signal spread reflected activation of perforant-path axons as well as recurrent excitation mediated by hilar mossy cells.
Figure 7, D–F, shows a similar experiment with hVOS 2.0 in the CA1 region where fluorescence was especially bright (Fig. 1C) and the largest hVOS signals were seen (Fig. 3D). Here, we displayed single-trial responses obtained without averaging. Stimulation in the sr evoked responses throughout the sr (Fig. 7D), and latency increased with distance (Fig. 7E). A sequence of color maps showed responses originating at the site of stimulation and sweeping through the CA1 region. These signals propagated more rapidly than in the DG, as expected for monosynaptic responses carried by Schaffer collaterals in the sr. Other sites were tested in slices from the hVOS 2.0 line. Generally, stimulation in the crest of the oml generated responses throughout the molecular layer, similar to that seen with hVOS 1.5 line 605 (Fig. 7, A–C). Stimulation in the iml, sg, and hilar region generated local responses that did not propagate as far (data not shown).
Figure 7 illustrated signal spread within a given region, but it is also of interest to study signal spread over larger areas and between regions. This can be accomplished by imaging responses in different regions to the same stimulation. Since the microscope was on a movable platform, we could change the field of view without moving the preparation or stimulation electrode. In this way, we prepared a composite of four regions encompassing the entire arc of the sp through the CA1, CA2, and CA3 regions of a slice from an hVOS 2.0 animal (Fig. 8, A–C). This enabled us to sample a large area while using our high-NA ×20 objective to record high-quality signals. Stimulation in the sr of the proximal CA3 region (near the DG, at location 1 in Fig. 8A) evoked responses that first appeared at the stimulation site and propagated throughout the regions imaged. Figure 8A shows the fluorescence traces superimposed on the corresponding images. A color map of maximum signal intensity shows that signals attenuated as they spread within the CA3 region but abruptly increased on entering the CA1 region at the CA2 border (Fig. 8B). There was a ∼17-ms latency for signal onset in the CA1 region ∼700 μm away (Fig. 8C). Similar results are shown in Fig. 8, D and E, for another site of stimulation. Stimulation in the sr of the CA3 region, but closer to CA1 region, again generated hVOS signals that spread into the CA1 region; strong signals were again seen in the CA1 region at CA2 border. This location had been identified as a “hot spot” in rat hippocampal slices using synthetic, voltage-sensitive dye (Chang et al. 2007). In both that study and in the present work, responses in the CA2 region were very small. Maximal responses of comparable amplitude were seen in both the CA3 and CA1 region at the CA2 border (Fig. 8F). The points in a plot of latency vs. distance fell close to a line, indicating propagation with a constant velocity, although there was more scatter in the CA2 region because with the small responses their latency could not be determined accurately. The slope of the latency vs. distance plot yielded a velocity of 41 mm/s (Fig. 8G).
Fig. 8.
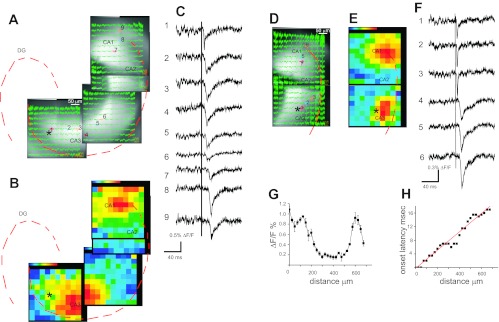
hVOS 2.0 signal spread from the CA3 to the CA1 region. A: images show 4 contiguous regions in which fluorescence changes were recorded in response to stimulation in the sr of the CA3 region close to h (site of stimulation indicated with asterisk). Responses in the CA3 region to stimulation at the indicated site were recorded in separate trials by moving the microscope while the stage and stimulating electrode were fixed. Fluorescence traces are superimposed on the images. The contours of the sp and sg are indicated with dashed red curves. B: the maximum signal intensity was encoded as color (red strongest, purple weakest) and mapped across the images shown in A with the sp and sg indicated as in A with dashed red curves. Largest responses were seen in the CA3 region and the CA1 region adjacent to the CA2 region. C: fluorescence traces from the locations indicated by the numbers in A. Response latency increased as the signals propagated from the CA3 region to the CA1 region. The vertical line indicates the time of stimulation. D: a similar experiment to that in A–C, but here the stimulus site was close to the CA2 region. Two contiguous regions are shown in which fluorescence changes were recorded in response to stimulation in the sr of the CA3 region. The sp contour is indicated with a red dashed curve. E: map of maximal responses at each location, as in B, shows largest responses near the site of stimulation and in the CA1 region adjacent to the CA2 region. F: traces from numbered locations indicated in D. G: the amplitude of the fluorescence change is plotted vs. distance from the stimulation site. As in F, responses are large in the CA3 and CA1 regions and small in the CA2 region. H: the onset latency increases linearly with distance from the site of stimulation. All traces are averages of 10 trials. Stimulation was 0.18 ms, 200 μA; [DPA] = 4 μM.
DISCUSSION
This study has demonstrated that hVOS probes genetically targeted to neurons in transgenic mice can be used to image electrical activity in brain slices. In transgenic lines expressing two different high-performance probes, we were able to record clear signals arising from populations of neurons. These signals depend on the presence of DPA to provide a voltage-sensitive FRET interaction with the membrane-targeted CeFP used in both of our hVOS probes. The fluorescence changes evoked by electrical stimulation had the expected dependence on excitatory synaptic receptors and voltage-gated Na+ channels and spread through slices in a manner consistent with known circuitry.
The thy-1 promoter targets neurons, and this can account for the differences between hVOS signals and the synthetic dye di-4-ANEPPDHQ (Fig. 6), which labels cells nonspecifically so that signals arise from both neurons and glia (Konnerth et al. 1987). Specificity between neurons and glia thus points the way toward more selective targeting and greater specificity of labeling and imaging. Because the targeting strategy used here made use of a general neuronal promoter (thy-1), the targeted neurons and their processes were closely packed so that signals from individual cells could not be resolved. Future efforts will be directed toward more specific targeting strategies to produce sparser labeling. Individual cells will be visible, and an attempt can be made to use hVOS to study their activity.
Among the hVOS transgenic lines reported here, different probe expression patterns were achieved. Thus, as reported previously with the thy-1.2 expression construct used here (Feng et al. 2000), lines generated with the same transgene exhibit different expression patterns. In the present study, probe expression was seen in granule cells and pyramidal cells in all 5 lines and in mossy cells in 3 out of 5 lines. Although thy-1-fluorescent protein transgenic mice exhibited an extremely wide range of expression patterns (Feng et al. 2000), the variation was across many structures. Within some structures, expression showed less variation. For example, 20 out of 25 lines showed expression in all motor axons, and 19 out of 25 showed expression in all cerebellar mossy cells. Other cell types were labeled with lower frequency. Our finding that 5 out 5 lines show dentate granule cell and hippocampal pyramidal cell expression and 3 out 5 show hilar mossy cell expression clearly falls within this spectrum. These results suggest that the thy-1.2 promoter can drive expression in these 3 types of neurons from different chromosomal locations. The ease of targeting mossy cells is fortunate considering how poorly understood they are (Henze and Buzsáki 2007).
The expression pattern of hVOS 2.0 indicates that this probe is preferentially expressed in axons (Figs. 1 and 2). Particularly strong expression was noted in the mossy fiber axons of the sl and the mossy cell axons of the iml in the DG. Although axonal expression was not noticed previously when this probe was expressed in cultured neurons (Wang et al. 2010), the GAP-43 motif incorporated into the NH2 terminus of this probe has been shown to sort proteins to axons (El-Husseini et al. 2001; Mosevitsky 2005). This observation raises the possibility of developing probes that target different subcellular compartments. It may seem surprising that despite the apparent preferential targeting of hVOS 2.0 to axons, the signals had the kinetic and pharmacological properties of postsynaptic responses (Fig. 5). However, activating excitatory synapses will still depolarize these cells, and axons have space constants of several hundred micrometers, allowing somatic depolarizations to spread to axons (Christie et al. 2012).
With the present success of hVOS imaging in transgenic mice, this work demonstrates the viability of studying electrical activity in genetically targeted cells in intact neural circuits. To study the functions of different types of cells, mice can be generated with hVOS probes under the control of various promoters such as the glutamic acid decarboxylase (GAD) promoter to drive expression in inhibitory GABAergic neurons (Lopez-Bendito et al. 2004; Oliva et al. 2000), the GFAP promoter to target glia cells (Zhuo et al. 1997), or the Hb9 promoter to target neurons that generate locomotor rhythms (Hinckley et al. 2005). A variety of strategies can be used to target probes to more precisely specified populations of neurons (Huang et al. 2010; Miyoshi and Fishell 2006). Targeting hVOS probes in this way will enable imaging experiments to reveal the circuit activity of these cells and allow investigators to study the relationships between the electrical activity of different members of these neuronal populations. Of particular interest in this regard are inhibitory interneurons, which are sparsely distributed and difficult to classify but play pivotal roles in most cortical functions (Freund and Buzsáki 1996; Markram et al. 2004). Interneurons form networks connected by electrical and chemical synapses (Connors and Long 2004) and contribute to an array of electrical oscillations with diverse spatial and temporal properties (Bartos et al. 2007; Klausberger and Somogyi 2008). The recent development of Cre driver lines that target different GABAergic neurons (Taniguchi et al. 2011) will make it possible to target hVOS probes to specific types of interneurons to study their network activity.
GRANTS
This work was funded by National Institute of Neurological Disorders and Stroke Grant NS-061150 and an American Recovery and Reinvestment Act (ARRA) supplement to this grant.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
D.W. and M.B.J. conception and design of research; D.W., S.M., and Z.Z. performed experiments; D.W., S.M., and M.B.J. analyzed data; D.W. and M.B.J. interpreted results of experiments; D.W. and M.B.J. prepared figures; D.W. and M.B.J. drafted manuscript; M.B.J. edited and revised manuscript; M.B.J. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Lea Ziskind-Conhaim, Baron Chanda, Samuel Pfaff, and Tiffany Poon for contributions to an earlier collaboration that led to the present study. We thank Peter Bayguinov for help in image analysis along with Logan Riemer and Paanteha Moghadam for comments on the manuscript. We thank other members of the Jackson laboratory for helpful discussion and Nima Ghitani for helpful insights arising from a parallel study of hVOS probes expressed by in utero electroporation. We thank Guoping Feng for providing the thy-1.2 expression construct and Kathy Krentz of the Wisconsin Transgenic Animal Facility for generating the mice used in this study.
REFERENCES
- Akemann W, Mutoh H, Perron A, Rossier J, Knopfel T. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat Methods 7: 643–649, 2010 [DOI] [PubMed] [Google Scholar]
- Amaral DG, Dolorfo C, Alvarez-Royo P. Organization of CA1 projections to the subiculum: a PHA-L analysis in the rat. Hippocampus 1: 415–435, 1991 [DOI] [PubMed] [Google Scholar]
- Amaral DG, Scharfman HE, Lavenex P. The dentate gyrus: fundamental neuroanatomical organization (dentate gyrus for dummies). Prog Brain Res 163: 3–22, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J 82: 509–516, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Lee H, Pieribone VA, Cohen LB, Isacoff EY, Knopfel T, Kosmidis EK. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J Neurosci Methods 161: 32–38, 2007 [DOI] [PubMed] [Google Scholar]
- Bartos M, Vida I, Jonas P. Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci 8: 45–56, 2007 [DOI] [PubMed] [Google Scholar]
- Bradley J, Luo R, Otis TS, DiGregorio DA. Submillisecond optical reporting of membrane potential in situ using a neuronal tracer dye. J Neurosci 29: 9197–9209, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canepari M, Zecevic D. (editors). Membrane Potential Imaging in the Nervous System. New York: Springer, 2010, p. 168 [Google Scholar]
- Chanda B, Blunck R, Faria LC, Schweizer FE, Mody I, Bezanilla F. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat Neurosci 8: 1619–1626, 2005 [DOI] [PubMed] [Google Scholar]
- Chang PY, Jackson MB. Heterogeneous spatial patterns of long-term potentiation in rat hippocampal slices. J Physiol 576: 427–443, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PY, Taylor PE, Jackson MB. Voltage imaging reveals the CA1 region at the CA2 border as a focus for epileptiform discharges and long-term potentiation in hippocampal slices. J Neurophysiol 98: 1309–1322, 2007 [DOI] [PubMed] [Google Scholar]
- Christie JM, Chiu DN, Jahr CE. Ca2+-dependent enhancement of release by subthreshold somatic depolarization. Nat Neurosci 14: 62–68, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LB, Salzberg BM. Optical measurement of membrane potential. Rev Physiol Biochem Pharmacol 83: 35–88, 1978 [DOI] [PubMed] [Google Scholar]
- Connors BW, Long MA. Electrical synapses in the mammalian brain. Annu Rev Neurosci 27: 393–418, 2004 [DOI] [PubMed] [Google Scholar]
- Dean C, Liu H, Dunning FM, Chang PY, Jackson MB, Chapman ER. Synaptotagmin-IV modulates synaptic function and long-term potentiation by regulating BDNF release. Nat Neurosci 12: 767–776, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D, He Y, Mutoh H, Baker BJ, Cohen L, Akemann W, Knopfel T. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS One 2: e440, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Craven SE, Brock SC, Bredt DS. Polarized targeting of peripheral membrane proteins in neurons. J Biol Chem 276: 44984–44992, 2001 [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28: 41–51, 2000 [DOI] [PubMed] [Google Scholar]
- Freund TF, Buzsáki G. Interneurons of the hippocampus. Hippocampus 6: 347–470, 1996 [DOI] [PubMed] [Google Scholar]
- Gonzalez JE, Tsien RY. Voltage sensing by fluorescence resonance energy transfer in single cells. Biophys J 69: 1272–1280, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinvald A, Frostig RD, Lieke E, Hildesheim R. Optical imaging of neuronal activity. Physiol Rev 68: 1285–1366, 1988 [DOI] [PubMed] [Google Scholar]
- Grinvald A, Manker A, Segal M. Visualization of the spread of electrical activity in rat hippocampal slices by voltage-sensitive optical probes. J Physiol 333: 269–291, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero G, Siegel MS, Roska B, Loots E, Isacoff EY. Tuning FlaSh: redesign of the dynamics, voltage range, and color of the genetically encoded optical sensor of membrane potential. Biophys J 83: 3607–3618, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze DA, Buzsáki G. Hilar mossy cells: functional identification and activity in vivo. Prog Brain Res 163: 199–216, 2007 [DOI] [PubMed] [Google Scholar]
- Hinckley CA, Hartley R, Wu L, Todd A, Ziskind-Conhaim L. Locomotor-like rhythms in a genetically distinct cluster of interneurons in the mammalian spinal cord. J Neurophysiol 93: 1439–1449, 2005 [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Taniguchi H, Miao H, Kuhlman S. Genetic labeling of neurons in mouse brain. In: Imaging in Developmental Biology: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2010, p. 199 [Google Scholar]
- Jackson MB, Scharfman HE. Positive feedback from hilar mossy cells to granule cells in the dentate gyrus revealed by voltage-sensitive dye and microelectrode recording. J Neurophysiol 76: 601–616, 1996 [DOI] [PubMed] [Google Scholar]
- Jin L, Mutoh H, Knopfel T, Cohen LB, Hughes T, Pieribone VA, Isacoff EY, Salzberg BM, Baker BJ. Genetically encoded protein sensors of membrane potential. In: Membrane Potential Imaging in the Nervous System, edited by Canepari M, Zecevic D. New York: Springer, 2010, p. 157–163 [Google Scholar]
- Jin W, Zhang RJ, Wu JY. Voltage-sensitive dye imaging of population neuronal activity in cortical tissue. J Neurosci Methods 115: 13–27, 2002 [DOI] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321: 53–57, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopfel T, Diez-Garcia J, Akemann W. Optical probing of neuronal circuit dynamics: genetically encoded versus classical fluorescent sensors. Trends Neurosci 29: 160–166, 2006 [DOI] [PubMed] [Google Scholar]
- Kojima S, Nakamura T, Nidaira T, Nakamura K, Ooashi N, Ito E, Watase K, Tanaka K, Wada K, Kudo Y, Miyakawa H. Optical detection of synaptically induced glutamate transport in hippocampal slices. J Neurosci 19: 2580–2588, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konnerth A, Obaid AL, Salzberg BM. Optical recording of electrical activity from parallel fibres and other cell types in skate cerebellar slices in vitro. J Physiol 393: 681–702, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods 9: 90–95, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bendito G, Sturgess K, Erdelyi F, Szabo G, Molnar Z, Paulsen O. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb Cortex 14: 1122–1133, 2004 [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci 5: 793–807, 2004 [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G. Directing neuron-specific transgene expression in the mouse CNS. Curr Opin Neurobiol 16: 577–584, 2006 [DOI] [PubMed] [Google Scholar]
- Mosevitsky MI. Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int Rev Cytol 245: 245–325, 2005 [DOI] [PubMed] [Google Scholar]
- Obaid AL, Loew LM, Wuskell JP, Salzberg BM. Novel naphthylstyryl-pyridium potentiometric dyes offer advantages for neural network analysis. J Neurosci Methods 134: 179–190, 2004 [DOI] [PubMed] [Google Scholar]
- Oliva AA, Jr, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci 20: 3354–3368, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai R, Repunte-Canonigo V, Raj CD, Knopfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci 13: 2314–2318, 2001 [DOI] [PubMed] [Google Scholar]
- Scharfman HE, Sollas AL, Smith KL, Jackson MB, Goodman JH. Structural and functional asymmetry in the normal and epileptic rat dentate gyrus. J Comp Neurol 454: 424–439, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron 19: 735–741, 1997 [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71: 995–1013, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods 5: 683–685, 2008 [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang Z, Chanda B, Jackson MB. Improved probes for hybrid voltage sensor imaging. Biophys J 99: 2355–2365, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Sun B, Zhang CL, Fine A, Chiu SY, Messing A. Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev Biol 187: 36–42, 1997 [DOI] [PubMed] [Google Scholar]


