Figure 1. Structure of HBeAg.
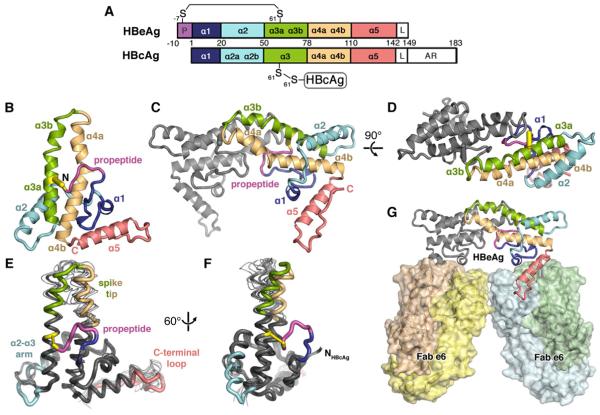
(A) Domain schematic of HBeAg and HBcAg, which share the core domain but differ in that HBeAg retains the N-terminal propeptide (P; magenta) and lacks the RNA-binding arginine-rich domain (AR). The linker region (L) is present, but disordered in crystal structures of both antigens.
(B) Ribbon diagram of HBeAg monomer, colored blue to red from N- to C-terminus, with the propeptide (magenta) shown forming an intramolecular disulfide (yellow) between C(-7) and C61. Secondary structure elements are entirely α-helical (α1-α5); propeptide is irregular coil.
(C, D) Ribbon diagram of the HBeAg dimer. viewed axially. Front subunit is colored according to the scheme in A; rear subunit is colored gray. Hairpins of the α3b and α4a helices from each subunit form the dimer interface, supported by the propeptides intercalated between them.
(E, F) Superposition of HBeAg and HBcAg monomers (thick and thin ribbon, respectively) (Packianathan et al., 2010; Wynne et al., 1999).
(G) Crystal asymmetric unit depicts HBeAg dimer complexed with two Fab e6 molecules (shown as molecular surface) binding at α5 and the C-terminal loop (red).
