Table 1.
Predicted and experimental binding affinity for compounds identified using Autodock 4.2 to dock compounds into human PMK. NMR and fluorescence techniques were then used to experimentally verify binding, and determine dissociation constants (Kd values).
| Chemical | Predicted Lowest Binding Energy (kcal/mol) |
Kd (µM) NMR [Residue used for data fitting] |
Kd (µM) Fluorescence |
|---|---|---|---|
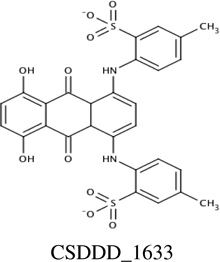 |
−11.6 | 55.4 ± 12.9 [G177] |
31 ± 8 |
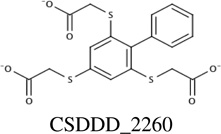 |
−10.34 | 6.3 ± 5.7 [T128] |
14.6 ± 4.6 |
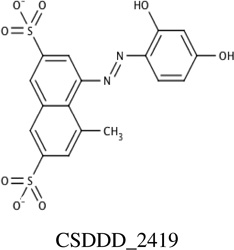 |
−11.8 | NDa [Q136] |
12 ± 2 |
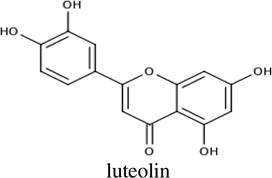 |
−9.2 | NDa [A172] |
61.0 ± 19.5 |
The NMR fit was not reliable because in addition to a specific binding event that seems to occur at low concentrations, there is also a non-specific effect that does not plateau.
