Fig. 2.
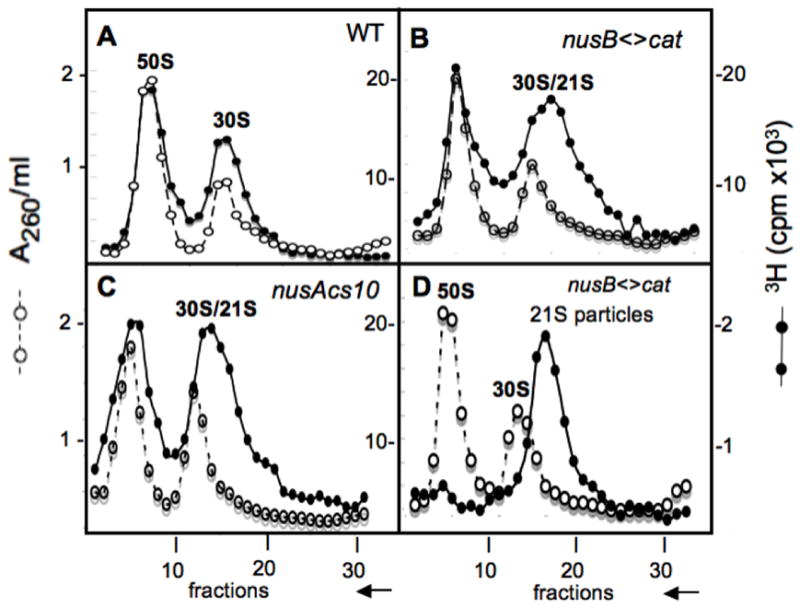
Identification of 21S ribosomal particles in NusA and NusB cold-sensitive mutants at 20°. Sucrose gradients were used to analyze ribosome subunits. The 50S and 30S subunits from wild-type bacteria serve as markers and were monitored in each fraction by A260 (left Y-axis). Ribosome subunits from cells whose RNA was labeled with [3H]-uridine for 1 hr at 20° before harvesting, monitored by [3H] (right Y-axis). Profiles of ribosome subunits prepared from the strain W3110 (panel A), the nusB< >cat derivative NB421 (panel B), and the nusAcs10 derivative NB755 (panel C) are shown as sedimenting from right to left as indicated by the arrow on the X-axis. Fraction 15 taken from the right shoulder of the 30S peak shown in panel B was mixed with unlabeled 50S and 30S subunits from a wild type strain and analyzed by sucrose gradient (see panel D), where a [3H]-21S peak is observed. Bacterial crude extracts were prepared and sedimented on sucrose gradients as described in Methods.
