Figure 4. Protein Folding of Selected DNA Glycosylases and Trp-Asp-Trp Triad in S. pombe Mag2 and E. coli AlkA.
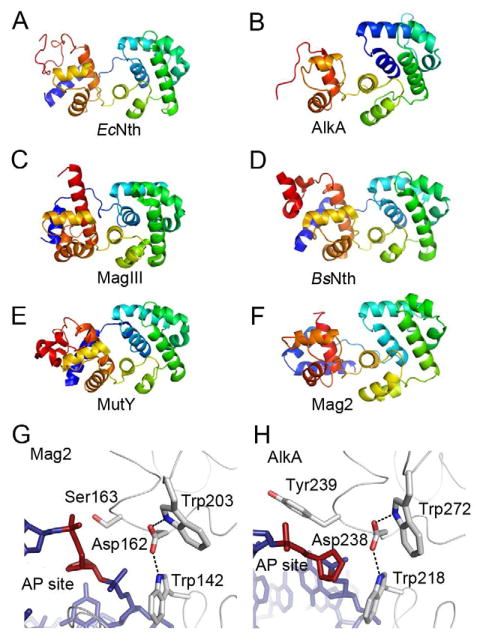
(A)–(F) Protein folding of (A) E. coli endonuclease III (Nth) (Thayer et al., 1995), (B) E. coli 3mA DNA glycosylase II (AlkA) (Hollis et al., 2000a), (C) H. pylori 3mA DNA glycosylase MagIII (Eichman et al., 2003), (D) B. stearothermophilus endonuclease III (Nth) (Fromme and Verdine, 2003), (E) B. stearothermophilus adenine DNA glycosylase (MutY) (Fromme et al., 2004), and (F) S. pombe Mag2. Color spectrum goes from N-terminal blue to C-terminal red. Only the C-terminal α-helical part of AlkA and the N-terminal part of B. stearothermophilus Nth are shown. (G) and (H) Close-up view of the AP site binding region in (G) S. pombe Mag2 and (H) E. coli AlkA (Hollis et al., 2000a) with stick models of the DNA (blue with abasic site in red). Both structures contain a Trp-Asp-Trp motif. The bulky side chain of Tyr239 in AlkA (H) pushes on the 5′ phosphate of the AP site as part of the flipping mechanism, while the less bulky Ser163 in Mag2 (G) allow the accommodation of a non-flipped AP site.
