Fig. 2. Amino acids affected by interaction suppressors.
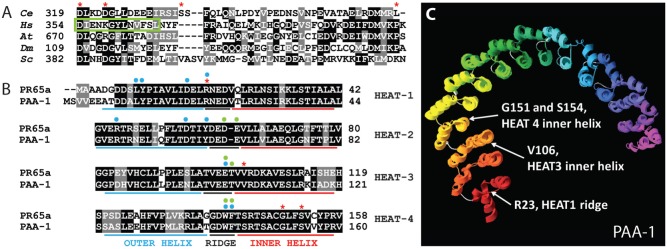
(A) Amino acid sequence alignment of RSA-1 and PP2A B″ regulatory subunits in H. sapiens (Hs) G5PR, D. melanogaster (Dm) Cg4733, S. cerevisiae (Sc) FRQ1, and A. thaliana (At) TON2. D319, D323, S335, and L367 intragenic suppressor locations are highlighted with red asterisks. The green box denotes the second of a tandem EF-hand motif that binds Ca2+ in some B″ subunits. (B) The first four N-terminal HEAT repeats of PAA-1 and human PR65. R23, V106, G151, and S154 suppressor locations are highlighted with red asterisks. Blue and green dots denote amino acids that physically interact (H-bonds or van der Waals) with a B (Strack et al., 2002; Xu et al., 2008) or B′ (Cho and Xu, 2007; Xu et al., 2006) regulatory subunit respectively. (C) C. elegans PAA-1 aligned to the published crystal structure of human PR65 (3dw8A). White arrows indicate suppressor locations. HEAT repeats are red at the N-terminus to purple at the C-terminus. Short loops between HEAT repeats are gray. See Table 1 for amino acid alterations in the suppressor mutations.
