FIG. 3.
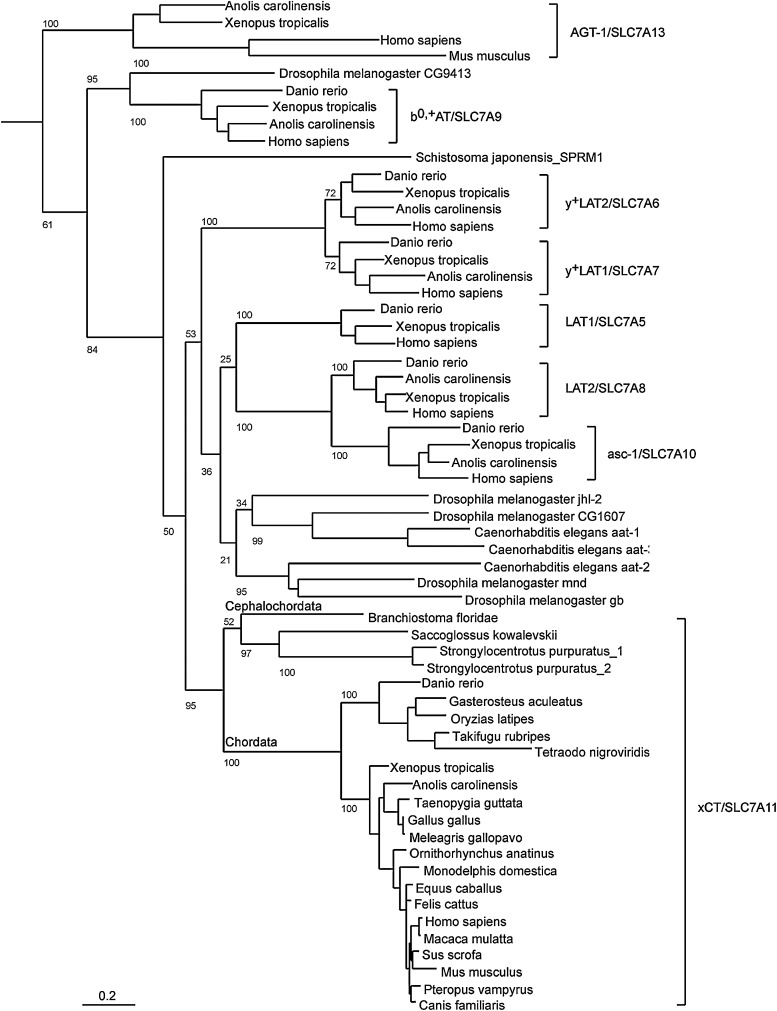
Phylogenetic analysis. Phylogenetic analysis of xCT/SLC7A11 orthologs and related heterodimeric amino acid transporter (HAT) proteins. xCT proteins from vertebrates, cephalochordata, hemichordata, and echinodermata; five putative Drosophila melanogaster LAT1/2 or xCT homologs; the three nonvertebrate HAT light-chain proteins from Caenorhabditis elegans and Schistosoma japonensis with a characterized function; and HAT orthologs from four evolutionarily separated vertebrates were included in the analysis. The distantly related SLC7A14 transporter was used as an outlier. Proteins were identified by BLAST search and used to generate multiple sequence alignments. The output was then used to generate a phylogenetic analysis using Maximum Likelihood and 100 bootstrap steps. The number of bootstrap iterations resulting in the branching shown is given as a number in front of each branch. The scale bar shows 0.2 (or 20%) sequence divergence (for methods see Supplementary Data [available online at www.liebertpub.com/ars]).
