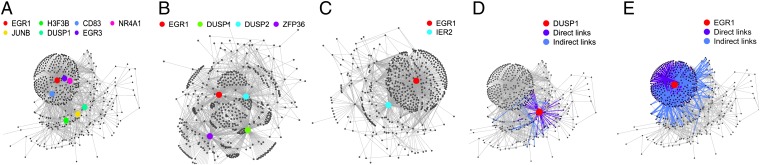
Fig. 2.
Visualization of inferred networks. The gene regulatory network of the most-aggressive leukemic B cells (A), the indolent leukemic B cells (B), and healthy B cells (C) are represented. Nodes represent genes, and edges statistical relationships between genes. For each network, hubs are highlighted in color. As the number of hubs decreases between aggressive, indolent, and healthy networks, the structure of the network is changed. Subnetworks for DUSP1 (D) and EGR1 (E) in the most-aggressive leukemic B-cell networks. The concerned gene is highlighted in red. Direct links are shown in navy blue, and indirect links are shown in pale blue. EGR1 is a gene whose influence is very large, because its subnetwork takes a large part of the complete network. In contrast, DUSP1 has a limited subnetwork. Visualization generated using R and R package igraph.