Fig. 3.
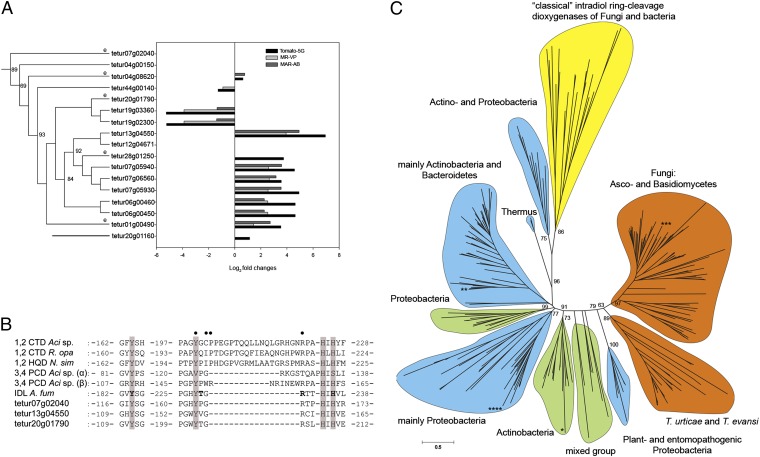
Intradiol ring-cleavage dioxygenases (ID-RCDs) in T. urticae. (A) Phylogenetic relationship of T. urticae ID-RCDs linked to expression levels (log2(FC)) in acaricide-multiresistant strains (MR-VP and MAR-AB) and after host plant shift to tomato (S. lycopersicum) for five generations. Genes with detected orthologs in T. evansi are depicted with an “e”. (B) Alignment of conserved residues in “classical” and secreted ID-RCDs. CTD, catechol ID-RCD; PCD, protocatechuate ID-RCD; HQD, hydroxyquinol ID-RCD; IDL, intradiol dioxygenase-like (cd03457); Aci, Acinetobacter sp.; R. opa, Rhodococcus opacus; N. sim, Nocardia simplex; A. fum, A. fumigatus. Tetur07g02040, tetur13g04550, and tetur20g01790 are ID-RCD representatives of T. urticae. The two His-2 Tyr nonheme iron (III) binding sites are indicated by shading. Residues defined by crystallographic (46–48) studies to have an influence on substrate interaction in classical ID-RCDs (CTD, PCD, and HQD) are indicated by solid circles. The predicted binding residues of epicatechin in the protein sequence of A. fumigatus are indicated in boldface type (69). (C) Maximun-likelihood unrooted tree of 17 ID-RCDs of T. urticae (and five T. evansi orthologs) with 232 bacterial and fungal sequences. Color codes indicate the percentage of secretion within the clade: yellow, not secreted; blue, <55% secreted; green, 55–85% secreted; and orange, >85% secreted. All members of the T. urticae clade are secreted and form a sister clade to fungal secreted dioxygenases, sharing a most recent common ancestor with plant and entomopathogenic Proteobacteria. The classical biochemically characterized ID-RCDs (CTD, PCD, and HQD) are not secreted and cluster together as an outgroup. The phylogenetic positions of the ID-RCD protein sequences of Naegleria gruberi (Protozoa), Shistosoma mansoni (Metazoa), P. infestans (oomycete), and Haloferax volcanii (Archaea) are indicated by *, **, ***, and ****, respectively.