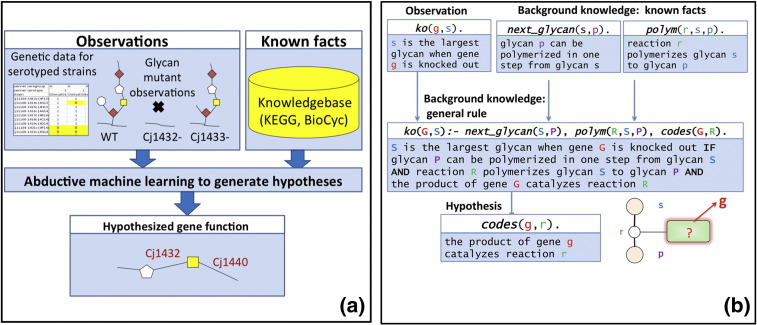
Fig. 1.
The logic-based machine learning approach to hypothesise functions for genes in the CPS synthetic pathway of C. jejuni. (a) Overview of machine learning. The background knowledge, consisting of known facts and rules, is used as a basis for abductive inference of novel hypothetical gene functions that might explain the experimental observations of single-knockout glycan phenotypes. (b) Simple system for abductive inference of gene functions in a metabolic network. Gene functions, represented by instances of the codes predicate, are hypothesised by the ILP program, based on the background knowledge of reactions, with the aim of explaining observed knockout phenotypes represented by the ko predicate. Capital letters G, R and S represent classes of object (genes, reactions, etc.) while small letters represent specific instances.