Figure 4.
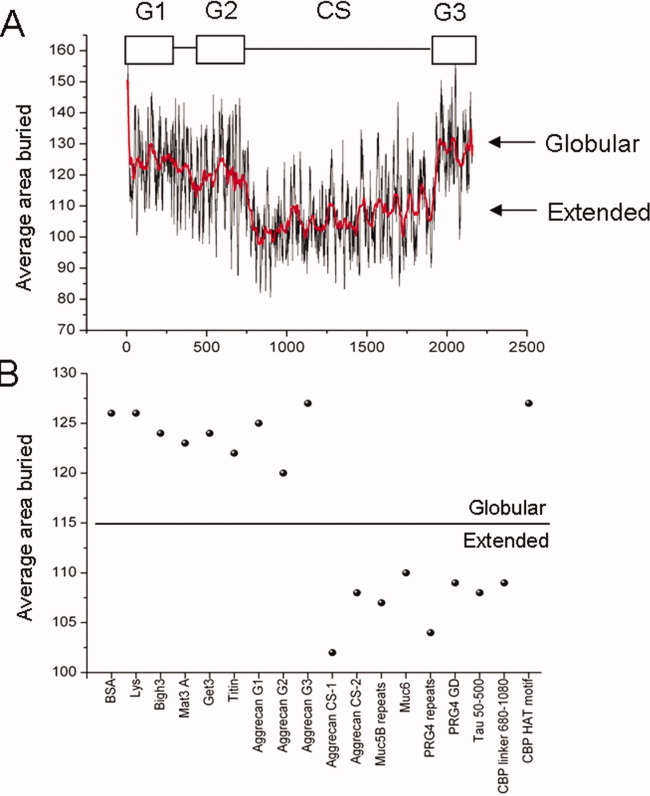
A: Bioinformatics analysis of the full sequence of aggrecan based on the average area buried upon folding for each amino acid. This simple algorithm developed by Rose et al.40 is able to highlight extended sequences. The domain architecture within aggrecan can clearly be distinguished as an overall decrease in the average area buried within the extended sequences corresponding to the chondroitin sulphate attachment (CS)-region. The red line is an 80-point Savkitzky–Golay average. By applying the same algorithm to other proteins and protein domains, there is a clear distinction in the score obtained between extended and globular characteristics (B). BSA, lysozyme (Lys), Bigh3, matrilin-3 A domain (Mat3 A), Get 3, titan, and the globular domains of aggrecan and the HAT motif of CBP, all produce an average domain score of >115, whilst the extended regions of aggrecan, muc5B tandem repeats, muc6, PRG4 tandem repeats and glycosaminoglycan attachment region (PRG4 GD), and the linker region of CBP, all produce scores <115. This analysis identifies a structural class distinction for some glycosylated and linker sequences and may correlate with stiffened and extended conformations.
