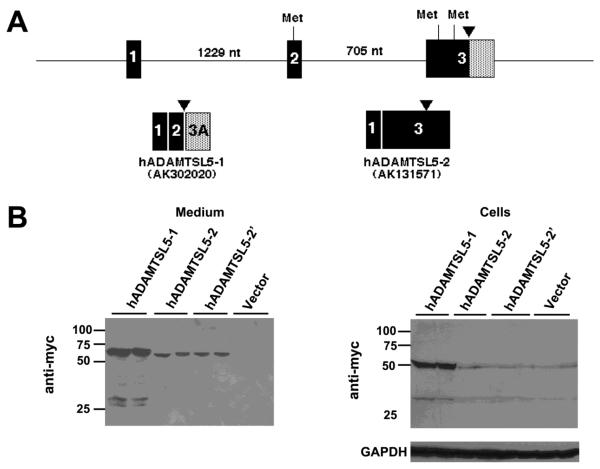
Figure 2. Alternative splicing at the 5′ end of ADAMTSL5.
A. Genomic region of the 5′ end of ADAMTSL5 (human gene) illustrating exons 1-3 and the intervening introns (size in nucleotides (nt)). The arrowhead indicates the location upstream of which the primary structures diverge (see Fig. 1B for sequences). Putative methionine codons (Met) within an acceptable Kozak consensus sequence are shown at the top of the figure. The pattern of exon splicing to generate the two alternatively spliced transcripts is shown below. Note that there are two potential exon 3 encoded start codons in the ADAMTSL5-2 transcript (Accession no. AK131571). B. Western blots of medium and lysate of HEK293F cells transfected with myc-His6 tagged ADAMTSL5-1 and ADAMTSL5-2 constructs. Lower levels of the expected 60 kDa protein species were obtained with ADAMTSL5-2 constructs in both cell lysate and medium. Abrogation of the upstream start codon in ADAMTSL5-2 (construct hADAMTSL5-2′) did not alter the protein level in cells or medium. The western blots show duplicate transfections for each construct.