Abstract
Antibiotic resistance mechanisms reported in Gram-negative bacteria are producing a worldwide health problem. The continuous dissemination of «multi-drug resistant» (MDR) bacteria drastically reduces the efficacy of our antibiotic “arsenal” and consequently increases the frequency of therapeutic failure. In MDR bacteria, the over-expression of efflux pumps that expel structurally-unrelated drugs contributes to the reduced susceptibility by decreasing the intracellular concentration of antibiotics. During the last decade, several clinical data indicate an increasing involvement of efflux pumps in the emergence and dissemination of resistant Gram-negative bacteria. It is necessary to clearly define the molecular, functional and genetic bases of the efflux pump in order to understand the translocation of antibiotic molecules through the efflux transporter. The recent investigation on the efflux pump AcrB at its structural and physiological level, including the identification of drug affinity sites and kinetic parameters for various antibiotics, may open the way to rationally develop an improved new generation of antibacterial agents as well as efflux inhibitors in order to efficiently combat efflux-based resistance mechanisms.
Keywords: Antibiotic resistance, RND family drug transporters, bacterial efflux pump, MDR bacteria, outer membrane permeability, selectivity, structure-function relationship
I. Introduction
Today, the treatment of bacterial infections is severely compromised by the emergence of bacteria that are resistant to multiple antibiotics. The dissemination of multi-drug resistant (MDR) Gram-negative bacteria thus drastically impairs the efficacy of antibiotic families and limits their clinical uses (Rice, 2006; Falagas & Bliziotis, 2007; Chopra et al., 2008; Blot et al., 2007; Gandhi et al., 2010). Mechanisms associated with modifications of membrane permeation processes, such as decreasing passive uptake (influx) or increasing active efflux of antibiotics, are now reported as key contributors of the bacterial MDR phenotype (Li & Nikaido, 2009; Nikaido, 2009; Piddock, 2006; Poole, 2007; Davin-Regli et al., 2008; Pagès et al., 2008)
To efficiently combat MDR bacteria, it is necessary to understand the molecular basis of the efflux mechanism which is involved in the limitation of intracellular (or periplasmic) concentration of all clinically used groups of antibiotics (Fig. 1). Indeed, the poly-specificity of bacterial efflux transporters, especially those belonging to resistance-nodulation-division (RND) family, (Li & Nikaido, 2009; Nikaido, 2009; Piddock, 2006; Poole, 2007) confers a resistance phenotype that can contribute to the acquisition of additional mechanisms of resistance including mutation of antibiotic targets (e.g. mutation in gyrase) or production of enzymes that degrade antibiotics (e.g. β-lactamases) and also reinforces the effects of these acquired mechanisms (Piddock, 2006; Poole, 2007; Davin-Regli et al., 2008). In addition, several reports mention an increase in the hospital dissemination of bacterial strains expressing a drug efflux mechanism (Li & Nikaido, 2009; Davin-Regli et al., 2008; Pagès et al., 2010) that is involved in the expulsion of various molecular families of antibacterial drugs and other chemicals. The structures of several individual components of the tripartite efflux systems, containing RND pumps, of Escherichia coli and Pseudomonas aeruginosa have now been resolved by X-ray crystallography and the models for the tripartite assembly has been presented (for reviews see Pos, 2009; Nikaido & Takatsuka, 2009; Murakami, 2008; Eicher et al., 2009; Yao et al., 2010).
Fig. 1.
Some efflux pump substrates.
Concerning the recognition and uptake of substrates, structural studies indicate that the transporters may capture their substrates directly from the periplasm or from the outer leaflet of the cytoplasmic membrane (Nikaido, 1996; Li & Nikaido, 2009). However, the molecular details of the process, at which level recognition and uptake occur, the kinetics, and the stoichiometry of the antiport process still require more work. The molecular features of energy transduction, such as proton antiport for AcrB and MexB, have to be clarified. These key steps of drug transport involve several and specific interplays between intramolecular and intermolecular conformational changes and intermolecular dynamics; they can be deciphered by using a combination of crystallography, modelling and molecular biology, energy calculation and biological-functional investigations. The deciphering of structure and function, e.g. structure-activity relationship (SAR), of the RND efflux pump and its individual components is required for an intelligent design of potential adjuvants to antibacterial agents for the therapy of MDR bacterial infections (Pagès & Amaral, 2009; Pagès et al., 2010). Efflux pumps such as AcrAB-TolC and MexAB-OprM, are essential for bacterial survival and colonization/virulence, especially during infection when the pathogen is attacked by noxious agents or colonizes host tissues (Piddock, 2007). AcrAB-TolC and the MexAB-OprM are the archetypes of RND-type drug efflux systems active in E. coli and P. aeruginosa clinical strains. In addition, there is a collection of genetically distinct RND efflux pump systems that can replace the main efflux pump system if it is deleted or deactivated (Li & Nikaido, 2009; Piddock, 2007; Poole, 2007). Moreover, data indicate that some drug transporters show a cooperative interaction or can act sequentially (Lee et al., 2000; Tal & Schuldiner, 2009).
A major problem with this drug transport is associated with the broad specificity of the efflux pump and it is quite difficult to discriminate between a “single” binding step and a correct recognition-binding inside the high-affinity pocket with an active conformation, allowing efficient translocation of the drug to the outer membrane channel. With the structural data, recent computer docking analyses have produced some informations (Takatsuka et al., 2010) but clearly required biological supports to improve possible models (Symmons et at., 2009; Schulz et al., 2010; Yao et al., 2010). This is a “key” point, not only regarding the clarification of the mechanistic and dynamics of the process but also regarding the clinical impact of this behaviour. The identification of the pump domains (amino acid residues) and the pharmacophoric groups available at the drug surface will be required to understand the common interaction taking place during the antibiotic transport.
The identification of potential targets in efflux systems cannot be achieved without a strong effort integrating information coming from different scientific endeavours. All details of interactions and processes that functionally govern and regulate the efficacy of the pump can be useful for the development of an effective new mode of therapy to combat MDR Gram-negative infections.
II. Efflux pump flexibility/versatility and clinical resistance: a key role for promiscuous transporters
A large collection of papers describing the involvement of multidrug resistance associated with efflux pump production in veterinary and animal fields has been published during the last decades. In this manuscript, we focus the presentation on the human clinical aspect and discuss, when it is possible, the relationships between patient treatment, antibiotic and efflux acting in the bacterial isolates.
1. Efflux and resistance in clinical Gram-negative isolates
It is now well-recognized that during antibiotic treatments of infected and/or colonized patients, several isolates, initially susceptible to different antibiotic classes, became less susceptible to a large number of structurally unrelated molecules (Fernández et al., 2011; Gwynn et al., 2010; Maragakis, 2010; Jones, 2010; Giamarellou & Poulakou, 2009; Rice, 2009; Wagenlehner et al., 2010). This observation concurs with the description of promiscuous/polyselective efflux pumps (of usually RND-type) able to recognize and expel a large panel of antimicrobial agents outside the bacterial cell including antibiotics, biocides, detergents, etc (Li & Nikaido, 2009; Poole, 2007; Piddock, 2006). Recent works clearly illustrates the role of efflux pump in fluoroquinolone resistant isolates of E. coli, an emerging resistant bacterium causing increasing health concerns (Yasufuku et al., 2011; Swick et al., 2011; Amábile-Cuevas et al., 2010). In addition, several authors report the involvement of efflux in resistant Salmonella isolates (Lunn et al., 2010; Smith et al., 2010; O’Regan et al., 2009). Two fluoroquinolone-resistant Shigella clinical isolates overexpressed the outer membrane channel TolC involved in the tripartite AcrAB-TolC efflux pump, in response to ciprofloxacin (Kim et al., 2008). Similarly, regarding the evolution of food-borne pathogens, the reduced susceptibility to macrolides in resistant Campylobacter isolates is associated with the presence of active efflux pumps combined or not with target mutations (Pérez-Boto et al., 2010; Mamelli et al., 2005).
It is important to mention that several unrelated antibiotic molecules, e.g. imipenem and fluoroquinolones, are able to select for the over-expression of efflux pumps in originally susceptible patient isolates (for reviews see Li & Nikaido, 2009; Davin-Regli et al., 2008; Poole, 2007). This indicates that the increased expression of efflux pump production also contribute to decreasing the intracellular antibiotic concentration, in addition to the selectivity of the efflux transporter. Moreover, in some isolates, efflux pumps are able to handle not only antibiotics but also biocides including disinfectants, antiseptics, sterilisants, preservatives that are frequently used in medical practice (Hegstad et al., 2011; Tumah, 2009; Li & Nikaido, 2009; Piddock, 2007;Poole 2007; Levy, 2002).
Today, a large number of enterobacterial strains isolated during an antibiotic therapy exhibit a multidrug resistant phenotype associated with the expression of an active AcrAB pumps (or AcrAB-like pumps) that have been characterized as a major mechanism involved in the reduction of antibiotic susceptibility. Several reviews described the role of AcrAB in the case of enterobacterial clinical isolates and demonstrated that the efflux pump, expressed in the resistant isolates selected during the patient treatment, recognized and efficiently expelled several drug molecules outside the bacterium (Davin-Regli et al., 2008; Li & Nikaido, 2009; Piddock, 2006; Poole, 2007). These data illustrate a bacterial strategy emerging under antibiotic pressure or in contact with chemicals exhibiting similar properties (e.g. structure, activity): this is mainly based on the over-expression of the polyspecific efflux pump, AcrAB-like, able to rapidly manage the intracellular concentration of a large panel of antimicrobial agents in enterobacterial isolates associated or not with a modification of the outer membrane permeability (e.g. alteration of lipopolysaccharide or down regulation of porins).
2. E. coli, E. aerogenes and K. pneumoniae efflux pumps and clinical resistance
The polyspecificity of AcrAB pump type is the core of the process that triggers the emergence and the spreading of efflux-producing bacteria during the treatment of Gram-negative bacterial infections. In addition, this pump expression reinforce the key role of outer membrane barrier, as the first bacterial defence line, in allowing the acquisition of additional mechanism of resistance (Davin-Regli et al., 2009; Li & Nikaido, 2009; Amaral et al., 2011). AcrAB-TolC overproduction affects the MICs of several antimicrobial agents, and to determine whether increased acrAB expression correlated with MDR, drug resistance data collected at hospitals were analyzed in the case of E. coli isolates resistant to fluoroquinolones (Becnel Boyd et al., 2009; Morgan-Linnell et al., 2009). Using a classification based on the level of MDR, depending on the number of antibiotic classes for which the tested isolate is not susceptible, Swick et al. (Swick et al., 2011) conclude that, in general, the more severe the MDR phenotype, the higher the probability that the isolate also overexpressed acrAB. Interestingly, no isolate classified as MDR was fluoroquinolone-susceptible suggesting the possibility of an underlying correlation between fluoroquinolone resistance and MDR. Because fluoroquinolones are heavily prescribed, there is strong selective pressure for bacteria to become resistant to them and a direct efflux has been demonstrated for many quinolones (Poole, 2007; Li & Nikaido, 2009). acrAB overexpression may be an interesting biomarker for MDR taking into account the mechanism by which the bacteria become MDR. (Swick et al., 2011).
The human gastrointestinal tract is a natural reservoir for E. coli, and this population is often associated with clinical infections. Consequently, during antibiotic treatment the selection and accumulation of quinolone-resistant strains likely occurs in this niche. In a recent work, Lautenbach and coworkers observed that about 50% of E. coli isolates exhibited an efflux pump overproduction (Lautenbach et al., 2010). This frequency is higher than that of prior reports (Komp Lindgren et al., 2003; Lautenbach et al., 2006), and suggests that efflux mechanism may be becoming more widespread over time. Similarly to E. coli, we have seen a serious increase in the prevalence of efflux-producing E. aerogenes strains isolated in a French hospital during the last decade (Chevalier et al., 2008). Moreover, it is interesting to note that despite the many recognized substrates of efflux pumps, the presence of organic solvent tolerance was associated with a greater likelihood of resistance to chloramphenicol but not to other antibiotics (Lautenbach et al., 2010).
The emergence of clinical isolates of E. aerogenes and K. pneumoniae exhibiting an over-expression of efflux pump, specifically AcrAB-TolC, has been reported by several teams (Bratu et al., 2009; Landman et al., 2009; Pagès et al., 2009; Bialek et al., 2010; Padilla et al., 2010; Davin-Regli et al., 2008). Interestingly, in a recent publication reporting the involvement of active efflux in clinical isolates of K. pneumoniae, 92% of tested isolates were able to expel 90% of penetrating ciprofloxacin (Aathithan & French, 2011). The most resistant isolates had multiple target site alterations and variable, but usually high, levels of efflux activity. High levels of efflux pump activity appeared to be the main or only fluoroquinolones (FQ) resistance mechanism in some isolates without target alterations (Aathithan & French, 2011). These MDR strains have been observed after antibiotic treament of colonized or infected patients and demonstrated a noticeable decrease of the susceptibility towards different antibiotic families. In some MDR isolates, the AcrAB overexpression is associated with a decreased expression of porins. Moreover, it has been demonstrated that the AcrAB-TolC efflux active in clinical isolates is able to recognize and pump fluoroquinolones, chloramphenicol, and tetracycline out of the bacterial cell (Malléa et al., 2003). It is clear that the combination of different mechanisms, e.g. impermeability/efflux/target mutation, is a very sophisticated and efficient response involved in these well-adapted isolates (Davin-Regli et al., 2008).
It is especially important to mention that the overproduction of this pump family represented by the AcrB-MexB archetype (Li & Nikaido, 2009; Piddock, 2006) is largely reported in clinical isolates collected during some antibiotic therapies. The effect of fluoroquinolone treatment has been reported to contribute to the emergence of efflux producers in E. coli (Swick et al., 2011). Interestingly, during an imipenem therapy clonally-related E. aerogenes MDR strains were isolated from the original susceptible isolate. These isolates are imipenem-resistant due to a porin deficiency and express active efflux pumps ejecting various unrelated antibiotics including quinolones, tetracycline and chloramphenicol. All strains overexpressed AcrA indicating the involvement of AcrAB pump in response to imipenem therapy (Bornet et al., 2000, 2003).
3. β-lactams and efflux pumps in resistant isolates
Regarding the resistance mechanisms, enzymes, impermeability, and efflux, which play a role in β-lactam susceptibility of Gram-negative bacteria, the role of efflux in clinical strains of Enterobacteriaceae was an issue that was much debated. It is only recently that the involvement of efflux pump in resistance against β-lactams has been strongly discussed for certain clinical enterobacterial strains, wheras such an involvement has been demonstrated in P. aeruginosa many years ago in a pioneering study (Li et al., 1994; Nikaido, 1996). The study of isolates exhibiting a resistance towards β-lactam must take into account various key biological parameters, the penetration/uptake and the role of porins (Nikaido, 2003; Pagès et al., 2008), the efflux, and the enzymatic production, e.g. β-lactamase (Bush & Jacoby, 2010). These various factors are often combined in clinical strains isolated after different therapies of infected patients. In addition, the influx and efflux can modulate the intracellular concentration of inhibitors of hydrolytic enzymes such as clavulanate or tazobactam, and consequently their action can be modified. Moreover, the activities of efflux pump blockers which are pump substrates are also dependent on the efflux pump expression (Pagès & Amaral, 2009; see section II.5). The roles of these intricate elements that are involved in the control of the internal concentration of antibacterial molecules are difficult to assess systematically in large panels of clinical isolates. Although the involvement of efflux pump activity in drug resistance has been clearly documented in K. pneumoniae clinical isolates with regard to various antibiotic families, including chloramphenicol, tetracyclines and quinolones, it has never been clearly established in the case of the β-lactam molecules (Hasdemir et al., 2004; Källman et al., 2008; Tran et al., 2009). However, such a mechanism is likely to be active in a group of K. pneumoniae clinical strains isolated in a French hospital microbiology service over the last ten years (Pagès et al., 2009; Bialek et al., 2010). This hypothesis was based on a paradoxical β-lactam-resistant phenotype (reduced susceptibility to cefoxitin and susceptibility to both amoxicillin+clavulanic acid and extended spectrum cephalosporins) associated with a conjoint chloramphenicol and quinolone resistance supported by efflux mechanism. Thus, the production of AcrAB-TolC, components of the major pump in the Enterobacteriaceae (Li & Nikaido, 2009) was detected in K. pneumoniae isolates which are clonally unrelated strains, belonging to different bla gene-based groups of K. pneumoniae and free of plasmid-mediated AmpC-type β-lactamases (Haeggman et al., 2004; Bialek et al., 2010). Some isolates exhibited an overproduction of AcrAB-TolC compared to the level observed in the susceptible strain as previously reported in a collection of Turkish multi-drug resistant isolates (Hasdemir et al., 2004). In addition, the expression of K. pneumoniae porins (OmpK35, OmpK36) was similar to that of strain ATCC 11296 except for one strain which exhibited a porin-deficient phenotype. Phenylalanyl arginyl β-naphtylamide (PAβN), a well-known efflux pump inhibitor (EPI) (Pagès & Amaral, 2009; Lomovskaya & Bostian, 2006), decreased noticeably the MICs for various antibiotics including chloramphenicol, nalidixic acid, ofloxacin, erythromycin and cloxacillin (Pagès et al., 2009). These antibiotics are well-known substrates of efflux pumps but this is the first sound evidence demonstrating that cloxacillin is an efflux pump substrate in clinical K. pneumoniae isolates (although this was known in E. coli/Salmonella (Nikaido et al., 1998) and that its activity was improved by PAβN addition. Moreover, in the presence of sub-inhibitory concentrations of cloxacillin with PAβN, the authors observed a significant restoration of some β-lactam activities that is reported in a competitive assay (an illustration is presented in Table 1). The restoring effect was observed with the different β-lactam molecules, especially with cefoxitin, amoxicillin, piperacillin and cefepime whereas no MIC decrease was observed with other antibiotic families such as quinolones, macrolides, chloramphenicol (Pagès et al., 2009).
Table 1.
Restoration of β-lactam susceptibility by efflux pump inhibitor and cloxacillin combination in K. pneumoniae.
| Strains | Porins | MIC (mg/L) | |||||
|---|---|---|---|---|---|---|---|
| FOX | FOX+EPI | CLX | CLX+EPI | FOX+CLX | FOX+CLX+EPI | ||
| ATCC | OmpK35, 36 | 8 | 8 | 1024 | 64 | 4 | 0.25 |
| KPBJ6 | no porin | 64 | 64 | 2048 | 128 | 32 | 8 |
| KPBJ7 | OmpK35, 36 | 64 | 64 | 2048 | 256 | 32 | 0.5 |
FOX, cefoxitin; CLX, cloxacillin; EPI, efflux pump inhibitor
MIC for cefoxitin was determined in the absence, in the presence of EPI, or in the presence of combination of EPI plus cloxacillin (at 1/5MIC). MIC for cloxacillin was determined in the absence or in the presence of EPI. (adapted from Pagès et al., 2009)
Some efflux pump inhibitors, e.g. PAβN, quinoline derivatives, exhibit various activity spectrum against different pumps belonging to the AcrAB family (including MexAB) or in combination with different molecules belonging to various antibiotic classes (see section II.5). These data support quite well the large flexibility in the pump site and the corresponding affinity present in clinical isolates producing the same efflux transporter group. Moreover, these suggest that the innate amoxicillin resistance observed in K. pneumoniae would be due to a combination of an amoxicillin efflux for a part and of an amoxicillin hydrolysis for the other part if the porin expression was not altered. These data give some light on the profile of β-lactam recognition by efflux transporters, AcrAB, active in clinical isolates of K. pneumoniae. Thus, it is clearly demonstrated that efflux plays a key role in the intracellular concentration and susceptibility of β-lactams in K. pneumoniae. This role has been largely underestimated if we take into account the involvement of efflux in the acquisition of additional mechanisms of resistance as shown in Salmonella sp. and Campylobacter sp. (Ricci et al., 2006; Yan et al., 2006; Quinn et al., 2006). It should be mentioned that similar observations have been obtained with E. aerogenes and E. cloacae (JP Lavigne et al, unpublished data; Pérez et al., 2007; Glatz et al., 2010). It is also noted that imipenem treatment is able to select resistant isolates exhibiting a step-by-step adaptative phenotype with first, an efflux overproduction, and second, an efflux overproduction associated with a down expression of porin (Bornet et al., 2000, 2003). These results may explain some previous reports describing a paradoxical β-lactam resistance without consistent identification of a specific enzyme production or demonstration of a porin alteration and evoking a possible role of efflux in resistant enterobacterial isolates (Landman et al., 2009) or possibly reflecting AmpC-activity based on interpretive reading of wider antibiogram data (Hornsey et al., 2010). In addition, this involvement of efflux pump in the reduced susceptibility towards β-lactams has been shown in the laboratory strains of E. coli (Mazzariol et al., 2000) and proposed in the case of E. coli resistant isolates displaying a paradoxical cefuroxime resistance (Källman et al., 2009).
Interestingly, the clinical relevance of efflux pump activity observed in resistant isolates and its involvement in the reduced susceptibility towards some β-lactams fits well with the kinetic analyses performed using the periplasmic hydrolysis of β-lactams (Lim & Nikaido, 2010). The affinity for a specific antibiotic, based on the presence or absence of pharmacophoric groups involved in the pump-drug recognition process, is a key parameter in the efficacy of efflux (Pagès et al. 2010). The kinetics and dynamics aspects of pump-substrate interactions are discussed in the next chapter (III. Pump selectivity: a molecular aspect).
4. Efflux pumps in P. aeruginosa and A. baumannii
Pseudomonas aeruginosa is a well-known opportunistic pathogen, which is involved in severe human infections and produces RND multidrug efflux pumps, like MexAB-OprM, MexCD-OprJ, MexEF-OprN and MexXY-OprM (Li & Nikaido, 2009; Piddock, 2006; Poole, 2007). These pumps are described in various strains and contribute strongly to a reduced susceptibility towards antibiotics because of the very low permeability of the outer membrane porin channel (Kerr & Snelling, 2009; Li & Nikaido, 2009; Poole, 2007; Kiser et al., 2010; Lister et al., 2009). Regarding the role of drug efflux transporters in resistant Pseudomonas aeruginosa clinical isolates, several investigations reported a clear involvement of MexAB-OprM and MexXY-OprM in the expulsion of various usual antibiotic families (Ziha-Zarifi et al., 1999; Boutoille et al., 2004; Sobel et al., 2005; Tomás et al., 2010; Rodríguez-Martínez et al., 2009; Islam et al., 2009). For some antibiotic molecule, preference has been noted for specific pump transporters and a certain adaptive expression associated with other resistance mechanisms in clinical strains has been reported (Hirsch & Tam, 2010; Zavascki et al., 2010; Lister et al., 2009; Dunham et al., 2010).
A hospital study reports the existence of a noticeable relationship between the emergence of P. aeruginosa strains overproducing MexXY-OprM and the use of aminoglycosides, fluoroquinolones and specific antipseudomonal cephalosporins during the patients’ treatment (Hocquet et al., 2006, 2007, 2008). It seems that MexXY-OprM may confer significant advantage under antibiotic pressure in the hospital setting and can actively participate to the efflux of structurally unrelated antibacterial molecules. A study from the same team reported a balance in the efflux pump expression (MexXY versus MexAB) in clinical strains recovered from cystic fibrosis patients (Vettoretti et al., 2009a, 2009b). A recent study has reported that clonal type A and type B resistant isolates of P. aeruginosa isolated from Cystic Fibrosis patients in Ontario (Canada) exhibit an MDR pattern that can be partially caused by an increased expression of antibiotic efflux systems (Beaudoin et al., 2010). It is important to mention that the overproduction of MexXY is often reported in isolates showing a resistance towards aminoglycosides and some β-lactams, indicating that this pump may exhibit a similar affinity for cefepime, ciprofloxacin and amikacin. Interestingly, in clinical isolates that overproduce efflux pumps, MexXY can discriminate between various cephalosporins, e.g. strong efflux of cefbipirole but almost no activity for ceftazidime, reflecting the limit of polyselectivity (Baum et al., 2009; Peña et al., 2009).
Similarly to Pseudomonas, Acinetobacter baumannii, which also produce porins of very low permeability (Vila et al., 2007; Sugawara E & Nikaido H, unpublished results), has proven to be an increasingly important and worrisome species in health care-associated infections. The drug-resistant phenotype of this bacterial pathogen and its unusual and unpredictable susceptibility patterns make the therapeutic choices difficult and risky (Fishbain & Peleg, 2010; Zavascki et al., 2010). In Acinetobacter, different RND-type active efflux systems, such as AdeABC, AdeDE, AdeFGH, and AdeIJK, have been described (Coyne et al., 2011, 2010; Chau et al., 2004; Damier-Piolle et al., 2008; Magnet et al., 2001; Pachon & Vila, 2009; Roca et al., 2011; Ruzin et al., 2010). These systems confer a reduced susceptibility to aminoglycosides, fluoroquinolones, erythromycin, tetracycline and chloramphenicol to the producing bacteria. The clinical significance of differences in antimicrobial-susceptibility pattern among different Acinetobacter genomic DNA groups has been demonstrated (Houang et al., 2003), and the importance of efflux systems in multidrug resistance in Acinetobacter is increasingly appreciated (Coyne et al., 2011). A study conducted by Chu et al. (2006) have demonstrated the prevalence of various pumps in different clinical isolates: AdeABC is an active efflux system specific for genomic group 2, whilst AdeDE and AdeXYZ are systems predominantly expressed in genomic group 3. In a recent study, Chiu et al. (2010) reported that resistance to ampicillin/sulbactam associated with a reduced susceptibility towards fluoroquinolones and aminoglycosides had significant correlation with overexpression of the Ade efflux pump in A. baumannii.
5. Molecules blocking efflux pump and effect on the susceptibility of clinical resistant strains
Faced with the continuous spreading of resistant bacteria and the absence of new compounds in the pipeline, specially for the treatment of Gram-negative infections, research efforts are underway to rejuvenate and revitalise old antibiotics (for recent articles see Jones, in News and Analysis, 2010; Silver, 2011). To enhance/recover their activities by impairing various resistance mechanisms active in clinical pathogens, old molecules can be used in combination with “adjuvants or chemosensitizers” (Bal et al., 2010; van Bambeke et al, 2010, Pagès et al., 2010; Fischbach & Walsh, 2009). In this context, the polyselectivity of drug efflux pump in Gram negative isolates is of particular concern for the development of efficient new coumponds termed efflux pump inhibitors (EPIs). This new group of antibacterial molecules has been developed to circumvent the bacterial antibiotic resistance by blocking the efflux activity and restoring a normal intracellular concentration for the used antibiotic (Pagès & Amaral, 2009; Lomovskaya & Bostian, 2006). Among this original class of molecules, some of them are reported as possible competitive inhibitor to the antibiotic translocation by AcrB or MexB efflux pumps (Pagès et al., 2010). Several reviews have described the development of EPIs (for two very recent reviews see Pagès et al., 2010; van Bambeke et al., 2010). We focus this section on the use of documented EPIs on resistant clinical isolates and the possible conclusions regarding the selectivity of the AcrB transporter family.
At this moment, only a few investigations have been carried out to evaluate the effect of various EPIs on the activity of specific classes of antibiotics by using different clinical isolates (e.g. E. coli, E. aerogenes, K. pneumoniae, P. aeruginosa, etc) expressing drug efflux pumps. The activity of EPIs depends on their intracellular (or periplasmic) concentration, and consequently the uptake (influx) of EPI through the outer membrane is a key step (Pagès & Amaral, 2009). In some studies, it is reported that, depending on the antibiotic class and the type of EPI used, various discrepancies on the final restored level of antibiotic activity or/and on the amount of EPI needed for restoration can be observed regarding the restoring level of antibiotic activity. Different comparative studies have been performed using quinoline derivatives and also PAβN as standard (Malléa et al., 2003; Chevalier et al., 2004; Ghisalberti et al., 2006). It has been reported that with a same bacterial strain, for instance a selected E. aerogenes, K. pneumoniae or E. coli isolate, the activity spectrum of a defined EPI is different regarding the antibiotic tested as competitive substrate for pump activity. (This phenomenon was known from the time the first EPI was reported: thus PAβN, which decreases drastically the levofloxacin MIC in MexAB-OprM-overproducing P. aeruginosa, showed very little effect on the MIC of carbenicillin (Lomovskaya et al., 2001). In a recent study, Chevalier and coworkers demonstrated that PAβN and quinazoline derivatives do not have the same enhancer effect on ciprofloxacin, sparfloxacin and erythromycin activity evaluated in a E. aerogenes strain overproducing AcrB, a similar difference is also noted with K. pneumoniae and P. aeruginosa strains for other antibiotics (Chevalier et al., 2010; Mahamoud et al., 2011). This could be associated with the respective affinity of the ligands, e.g. EPI or antibiotic molecule, for the pump site, but also with the level of expression of the acting pump in the tested conditions. Moreover, some EPIs can be more active on a specific efflux pump: for example, there are EPIs that inhibit only the MexAB-OprM system among several RND pumps that contribute drug resistance to P. aeruginosa (Nakayama et al., 2003; also for a review see Pagès & Amaral, 2009). Regarding this point, it is important to mention that the original screening protocols used to develop and select EPIs, in terms of bacterial efflux target and antibiotic used as substrate, play an important role for the affinity and activity spectrum of EPI (Pagès & Amaral, 2009). This is illustrated in the case of the two EPI families (peptidomimetics and quinoline derivatives) recently developed (Pagès et al., 2010).
Similar comparison studies have been performed between PAβN and 1-(1-naphthylmethyl)-piperazine (NMP), an original EPI (Bohnert & Kern, 2005). The activity of NMP was different from that of PAβN on a collection of clinical isolates of E. coli, in particular regarding the macrolide resistance reversal (Kern et al., 2006). Moreover, NMP displays a moderate activity in reversing MDR in C. freundii, E. aerogenes, S. marcescens and K. pneumoniae clinical isolates. Its effects on the reversal of resistance depend on bacterial species and drug, and are different from those seen with PAβN (Schumacher et al., 2006). Thus the selectivity/efficacy of efflux pump and the activity of the respective EPIs on the degree of altered resistance are strongly interconnected. The question remains about the development of molecules that mimic the structure of a specific antibiotic molecule (via the use of appropriate pharmacophoric groups) to favor a directed improvement of the activity on a single antibiotic class. This may be a key question for the development and selection of future “adjuvants or chemosensitizers” able to restore a significant antibiotic concentration inside the bacterium.
III. Pump selectivity: a molecular aspect
1. Identity of affinity sites
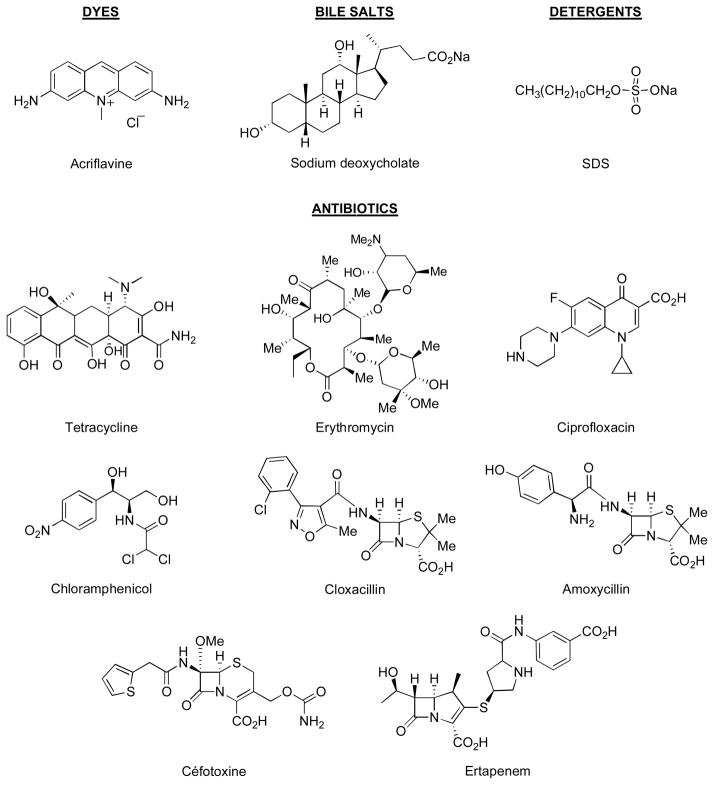
Our initial knowledge on the ligand-binding sites in RND pumps came from crystallographic studies. Thus very soon after the initial elucidation of the symmetric trimer structure of AcrB (Murakami et al., 2002), co-crystallization of AcrB with a few ligands (rhodamine 6G, ethidium, dequalinium, and ciprofloxacin) resulted in the electron density close to the ceiling of the central cavity in the transmembrane domain of AcrB (Yu et al., 2003). Further work with a N109A point mutant of AcrB showed additional binding of several ligands also to the entrance of large external cleft in the periplasmic domain (Yu et al., 2005)(see the green surface in Fig. 2B and 2C). This finding was later supported by the cocrystallization of deoxycholate at the same periplasmic site (Drew et al., 2008), as well as the unintended cocrystallization of ampicillin in the same site (Tornroth-Horsefield et al., 2007).
Fig. 2. Ligand-binding sites in the Binding Protomer of AcrB.
A. The “deep” binding site for minocycline and doxorubicin found by Murakami and coworkers (Murakami et al., 2006). The orientation of the trimeric protein is shown in the inset, in which each protomer is shown in different color (Access, mauve; Binding, sand; and Extrusion, green). The binding pocket is shown as an orange-colored surface, with the bound minocycline molecule in stick representation. The site consists of two subpockets, the narrow “Groove” and the much wider “Cave.” The amino acid residues shown in the surface representation were specified earlier (Takatsuka et al., 2010). Parts of the protein close to the viewer have been clipped away to show the site more clearly. B. The two plausible binding sites of AcrB. This shows the “deep”, “ultimate” binding site shown in A (as orange surface), as well as the putative site on the surface of the periplasmic domain, at the entrance of the large cleft (as green surface). The residues used to generate the latter surface are residues 566, 664, 666, 668, 673, 676, 717, and 828, identified by Husain and Nikaido (2010). To get this view, the model was rotated by 70° as shown at the bottom. No clipping was applied here. C. A possible path for the substrate, from the surface site (green surface) to the deep site (orange surface), with the tunnel shown in gray. This is a view from the top, and the presumed direction of the substrate flow is shown by an arrowhead. The tunnel was detected by the program Caver (www.caver.cz), and all figures were generated by using Pymol (www.pymol.org).
It was unclear, however, whether these ligand binding sites actually participated in the movement of ligands from the periplasm (or the membrane interior) into the TolC channel, and eventually into the outside medium. A major advance was made when three laboratories published asymmetric crystal structures of AcrB in 2006–2007 (Murakami et al., 2006; Seeger et al., 2006; Sennhauser et al., 2007). In these structures, each protomer takes a different conformation that appears to correspond to a different stage in the binding and extrusion of the ligand. Thus in this context, it was most important that Murakami and coworkers (Murakami et al., 2006) found two ligands, minocycline and doxorubicin, bound to a novel binding site deep in the interior of the periplasmic domain of one of the protomers (“the binding (or T) protomer”), a site nevertheless connected by a tunnel to the external cleft mentioned above (see the orange surface in Fig. 2, A, B, and C). With minocycline, the identification of the electron density was further strengthened by the use of its brominated derivative. The observation that this binding cavity collapses when the protomer is transformed into the next stage (“the extrusion (or O) protomer”) confirms that this site indeed occupies the central position in the movement of ligands. (The third protomer, “the access” (or L) protomer, might first interact with the free ligand in the periplasm). Finally, site-directed mutagenesis of residues lining this binding pocket indeed produces significant changes in the substrate specificity (Bohnert et al., 2008).
What is the significance of other binding sites? We initially thought that the central cavity was important, because site-directed mutagenesis there appeared to decrease the pumping activity (Yu et al., 2003). However, this was a laboratory artifact of the assay (Yu et al., 2005). Furthermore, the substrate specificity of AcrB was shown to depend entirely on the periplasmic domain through the domain exchange experiment with AcrD (Elkins & Nikaido, 2002), and so far there is no strong evidence suggesting a major role of this potential binding site. The situation, however, is different with the site in the external cleft (see the green surface in Fig. 2B). In addition to the crystallographic data already mentioned, we found that site-directed mutagenesis of some of the residues within the cleft decreased the pumping activity of AcrB substantially (Yu et al., 2005). Also some of the residues here were identified as residues whose mutations expanded the substrate range of P. aeruginosa MexD pump in the pioneering study by the Lomovskaya group (Mao et al., 2002). Finally, many of the residues in the cleft were shown to interact with the ligands that are being extruded, in our recent study of a wide range of AcrB residues using covalent modification of Cys mutant proteins by a rather lipophilic agent, BODIPY FL N(2-aminoethyl)maleimide (Husain & Nikaido, 2010). We therefore believe that most ligands are captured by the periplasmic domain of AcrB and its relatives first by binding to the entrance (or the bottom) of the external cleft, and then by entering the deep binding pocket identified by Murakami and others (Murakami et al., 2006)(Fig. 2C). We emphasize that the presumed path of substrate travel is all within the periplasmic domain, although some residues within the path are located close to the outer surface of the membrane bilayer. Thus in our current hypothesis, the transmembrane domain functions solely for proton translocation, which is coupled to the conformational changes in the periplasmic domain that propels drug extrusion (Nikaido & Takatsuka, 2009).
2. Substrate binding and affinity of the deep binding pockets of AcrB and MexB
We may be able to obtain some ideas of the affinity of the ligands to this most critical binding site, if we knew the kinetic constants of the RND pumps. However, it has been difficult to obtain these constants. In 2009, we have succeeded in obtaining Km and Vmax values for the efflux of cephalosporins by AcrB (Nagano & Nikaido, 2009) by determining the periplasmic concentrations of these compounds from their hydrolysis rates by periplasmic β-lactamase in intact cells. The rate of efflux is then calculated as the difference from the calculated influx across OM (that is proportional to the concentration difference across OM and to the permeability coefficient) and the measured hydrolysis rate. The Km values (or K0.5 values because with most cephalosporins the efflux rate vs. periplasmic concentration plots showed significant positive cooperativity) covered a wide range, from 5 μM (nitrocefin), 20 μM (cefamandole)(shown as an example in Fig. 3), 90 μM (cephalothin) to 290 μM (cephaloridine). In contrast, with penicillins examined (Lim & Nikaido, 2010), i.e. dicloxacillin, cloxacillin, oxacillin, azlocillin, mezlocillin, piperacillin, penicillin V, and ampicillin, the K0.5 values did not vary much, and were in the range of 0.7 to 1.1 μM. These values of Km or K0.5 are obviously not the KD values at the binding site, yet give some glimpse of them, that is, the affinity is probably not so strong as to give the KD values in nanomolar range, yet probably significant so that K0.5 values in the micromolar range become possible.
Fig. 3. Efflux and hydrolysis of cefamandole.
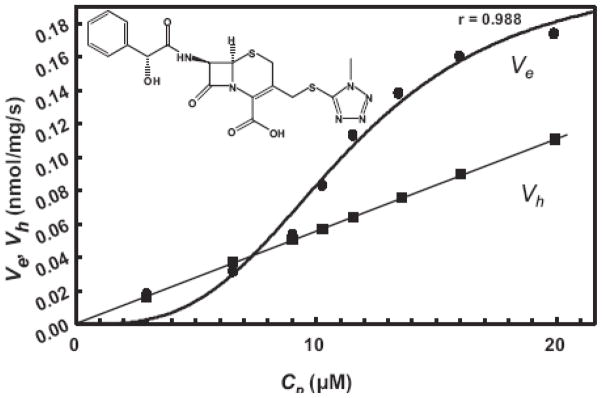
Efflux (Ve) and periplasmic hydrolysis rate (Vh) of cefamandole in E. coli strain HN1160, at different external concentrations (Co) of the drug. From (Nagano & Nikaido, 2009).
If the drugs bound only to the ultimate binding pocket of AcrB, then a more straightforward approach for the measurement of drug binding would become possible. Such an attempt was made with fluorescent ligands and isolated AcrB protein by using fluorescence polarization, and resulted in apparent KD values in the range of 5.5 to 74 μM (Su & Yu, 2007). However, it is doubtful if the workers observed the ligand binding to the true, deep binding pocket, because the binding was not much altered in the proton translocation mutants of AcrB, in which this pocket is nearly completely collapsed (Y. Takatsuka and H. Nikaido, unpublished observation).
Because the efforts to obtain AcrB crystals with substrates in their deep binding pockets were successful only with minocycline and doxorubicin (Murakami et al., 2006) in spite of extensive efforts in several laboratories, we decided to adopt the computer docking approach (Takatsuka et al., 2010). By using the apoprotein from the structure containing minocycline and asymmetric AcrB (PDB code 2DRD) and using the program Autodock Vina (Trott & Olson, 2010), we could show not only that minocycline is indeed predicted to become bound to the binding pocket as seen in the crystal structure, but also that many other ligands known to be extruded by the AcrB pump are predicted to be bound to this pocket with the calculated binding energies of between −7.3 and −12.0 kcal/mol. When the binding of known ligands were compared with that of non-substrates of AcrB (using this time a different AcrB model (Sennhauser et al., 2007) (PDB code 2J8S), the non-substrates (arabinose, lactose, kanamycin A) bound with significantly weaker binding energy (between −4.7 and −5.0 kcal/mol) than the known substrates that all bound with the energy calculated to be stronger than −7.4 kcal (H. Nikaido, unpublished results). Now that an asymmetric structure of another RND pump, MexB, is known in a form with the bound detergent, dodecylmaltoside (Sennhauser et al., 2009), we can compare the binding sites of AcrB and MexB. From the comparison of AcrAB-TolC and MexAB-OprM systems both expressed in the same E. coli host (Tikhonova et al., 2002), we note that AcrAB is much more efficient in pumping out macrolides oleandomycin and erythromycin, whereas MexAB excels in the extrusion of cinoxacin, a quinolone compound. Docking study with PDB models 2J8S and 2V50, however, did not produce data that fit with this difference in efficiency. Thus although erythromycin bound more strongly to AcrB (−9.8 kcal/mol) than to MexB (−8.7 kcal/mol), the binding of oleandomycin was predicted to be similar (−8.5 vs. −9.0 kcal/mol, respectively), and the expected preferential binding of cinoxacin to MexB was not apparent (−7.9 and −7.5 kcal/mol to AcrB and MexB). This could be due to the fact that the binding pockets are extremely flexible, consisting nearly entirely of loop regions, so that upon binding of different ligands the pocket itself undergoes a significant conformational change, which is not taken into account in the docking process that treats the protein as an inflexible structure. This interpretation was indeed supported by the finding that if the side-chains of five Phe residues (178, 610, 615, 617, and 628) and one Tyr residue (327) in the binding pocket are made flexible during docking, the MexB structure now binds cinoxacin better than the AcrB structure (H. Nikaido, unpublished results). Nevertheless, this is not a complete solution because the backbones are not moved during docking. Another possible explanation for this discrepancy between the drug extrusion efficiency and the binding affinity to the ultimate binding pocket might be the differences in the drug interaction with the earlier binding site at the cleft entrance, or with other parts the drug translocation channel.
One unexpected observation of the docking study with AcrB was that some ligands (e.g. minocycline, tetracycline, doxorubicin, novobiocin, nitrocefin, rhodamine 6G) were predicted to bind exclusively to the narrower “groove” portion of the pocket farther away from the membrane surface, whereas others (e.g. ethidium, carbenicillin, chloramphenicol, 1-(1-naphthylmethyl)piperazine [NMP]) binds nearly exclusively to the wider “cave” area of this large pocket (Takatsuka et al., 2010)(Fig. 2A). In order to test if there are indeed these two different binding modes in functioning AcrB, we carried out two lines of experiments. First, we tested whether the efflux of nitrocefin, a groove binder, is inhibited (presumably competitively) by the simultaneous presence of other ligands. The results were very clear. Although minocycline, a typical groove binder, inhibited the nitrocefin efflux very strongly, chloramphenicol, a cave binder, showed no evidence of competition even at rather high concentrations (Takatsuka et al., 2010). Second, we tested the covalent modification, by fluorescein maleimide, of Phe615Cys residue in the binding pocket of the binding protomer in the covalently linked AcrB trimer (Takatsuka & Nikaido, 2009). Because fluorescein maleimide is large and relatively lipophilic, we assume that the covalent modification involves the initial non-covalent binding of this reagent to the binding site, predicted to occur in the groove according to the docking study. Indeed, this covalent modification is inhibited strongly by minocycline and nitrocefin, both groove binders, but not at all by NMP, a cave binder (Takatsuka et al., 2010).
This concept of multiple subdomains of different specificity within a single large binding pocket is not entirely new. Thus Schumacher and coworkers (Schumacher et al., 2004) showed that within the large binding pocket of the efflux regulator protein QacR, two different ligand molecules may bind to different parts of the cavity. However, this notion is important for AcrB and its homologs because it may provide molecular level explanations for some biochemical observations that remained unexplained. For example, it has been a puzzle that the very effective inhibitor of RND-type efflux pumps, Phe-Arg-β-Naphthylamide prevents the efflux of fluoroquinolones very effectively from MexAB-OprM-expressing P. aeruginosa, while it showed barely detectable activity for the efflux of carbenicillin (Lomovskaya et al., 2001). Our docking studies with AcrB (Takatsuka et al., 2010) suggested that the preferred positions of binding of Phe-Arg-β-Naphthylamide and fluoroquinolones were similar, but carbenicillin bound to very different positions in the large binding pocket.
Recently, this notion of differential binding to subdomains of the binding pocket has received a strong support from the study of AcrAB-TolC overproducing clinical strains of Klebsiella pneumoniae (Pagès et al., 2009). Thus the addition of the inhibitor Phe-Arg-β-Naphthylamide decreased drastically the elevated MIC values of these strains for fluoroquinolones, macrolides, and chloramphenicol, whereas only modest decreases were observed for the MICs of many β-lactams. However, when subinhibitory concentrations of cloxacillin is added on top of Phe-Arg-β-Naphthylamide, then a spectacular decrease in MIC is observed for cefoxitin, piperacillin, amoxycillin, and cefepime. Since these strains produced only the endogenous, chromosomal SHV-1 type β-lactamase, which shows poor affinity to cloxacillin (Amicosante et al., 1988), this effect of cloxacillin cannot be explained by the inhibition of hydrolysis. Thus these studies show that a group of compounds, including Phe-Arg-β-Naphthylamide, macrolides, chloramphenicol, and fluoroquinolones, bind to one subdomain of the binding site and interfere with the efflux of each other, while some β-lactams presumably bind to another subsite preferentially, so that it takes another β-lactam, cloxacillin, to interfere with their efflux in an effective manner. (Cloxacillin in the presence of Phe-Arg-β-Naphthylamide did not further decrease the MICs of chloramphenicol, etc. However, this may be because their MICs were already very low without cloxacillin).
To summarize our current knowledge in this area, we learned from asymmetric crystal structures of AcrB and MexB containing ligands (Murakami et al., 2006; Sennhauser et al., 2009) how minocycline, doxorubicin, and dodecylmaltoside bind to the deep binding pockets in the periplasmic domains of these proteins. This appears to be the ultimate binding site for these pumps, as the pocket collapses in the next stage of the pump functional cycle, i.e. in the extrusion protomer. For other substrates of the pumps, computer docking appears to give reasonable predictions on their binding (Takatsuka et al., 2010), although the detailed parameters of the binding are rather unreliable especially because the flexible pocket is held as a rigid structure during the docking simulation. This study showed how different compounds could bind to different portions of this large site. This notion is supported by a striking observation on AcrAB-overproducing clinical isolates of K. pneumoniae (Pagès et al., 2009). One should not imagine, however, that binding to this ultimate binding pocket is the only critical factor in ligand efflux.
IV. The Effect of Pump Activity in Intact Cells
In the preceding section, we discussed the activity and specificity of RND multidrug efflux pumps often exhibiting an exceptionally wide substrate specificity. However, RND pumps have a construction spanning both the outer and inner membrane, and their effectiveness in increasing resistance levels is intimately connected with the presence of the OM permeability barrier. Thus the destruction of the OM permeability barrier for lipophilic drugs (by the use of polymyxin B nonapeptide (PMBN) (Vaara, 1992) decreased MIC values as much as the inactivation of the major RND pump genes in P. aeruginosa (Nikaido, 1998). It is clear, therefore, that discussion of the pump activity alone does not lead to the complete understanding of pump-mediated drug resistance, and that consideration of the interaction of this activity with the OM barrier becomes absolutely necessary.
1. How various drugs traverse the OM
Here we briefly summarize our current knowledge on how different groups of antibiotics penetrate across the outer membrane (see Table 2). β-Lactams penetrate mainly through the porin channels, as their carboxyl groups are rather strong acids (that of benzylpenicillin, as an example, has a pKa of 2.75 (Tsuji et al., 1977), and very little of these compounds exists in an uncharged protonated form. This is supported experimentally by the increased MIC in porinless mutants and the lack of sensitization by the perturbation of OM bilayer (Table 2). Nevertheless, lipophilic β-lactams diffuse across the lipid bilayers of the cytoplasmic membrane with measurable rates (Li et al., 1994). However, because the asymmetric lipid bilayer of OM is much less permeable than the bilayers of the conventional membranes (Plesiat & Nikaido, 1992), we can usually neglect their penetration through the bilayer region of OM. (Exceptionally, a lipophilic penicillin cloxacillin, which penetrates very poorly through porin channels (see below), becomes more effective when the OM bilayer is perturbed (Table 2). However, the diffusion across the unperturbed OM bilayer is likely to be very slow.) In penetrating through porin channel, the presence of lipophilic groups greatly retards the process (Nikaido & Rosenberg, 1983; Nikaido et al., 1983), presumably because the water molecules within the channel are highly ordered, and lipophilic drugs must perturb this ordered water structure (Kreusch et al., 1994). Negative charges tend to slow down the permeation strongly, in part because of the presence of the interior-negative Donnan potential across OM (Yoshimura & Nikaido, 1985). Zwitterionic compounds have an advantage that they do not suffer from this problem.
Table 2.
Outer membrane diffusion pathways and the role of AcrAB-TolC pump in susceptibility to various antibiotics in E. coli.
| Antibiotic | Main diffusion pathway across OM | Permeability Coefficient (nm/s) | Increase in Resistance (fold) (ΔompF) | Sensitization (fold) (Bilayer perturbation by PMBN) | Sensitization (fold) (ΔacrAB) |
|---|---|---|---|---|---|
| Penicillin G | Porin | 20 | 4 | 1–3 | 2 |
| Ampicillin | Porin | 980 | 2 | 1 | 2 |
| Carbenicillin | Porin | 7.5 | 16 | 3 | 4b |
| Cloxacillin | Porin | ~20 | NDa | 5–30 | 128 |
| Cefazolin | Porin | 600 | 2 | ND | 1 |
| Tetracycline | Porin | 100 | 1.5 | 1 | 2–4 |
| Chloramphenicol | Porin | ND | 2.5 | ND | 2–8 |
| Novobiocin | Bilayer | ND | ND | 10–100 | 32,64b |
| Erythromycin | Bilayer | ND | ND | 8–30 | 16,64b |
Not determined.
These are values determined by using Salmonella enterica serovar Typhimurium (Nikaido et al., 1998).
PMBN, polymyxin B nonapeptide.
The E. coli data are from Pugsley & Schnaitman (1978), Harder et al. (1981), Nikaido et al. (1983), Nikaido & Normark (1987), Vaara (1992), Thanassi et al. (1995), and Mazzariol et al. (2000).
Aminoglycosides are very hydrophilic, and polycationic. Thus we can assume that they permeate through the porin channel, although we still lack hard evidence on this point. Although a paper exists (Nakae & Nakae, 1982) claiming this has been experimentally established, this study uses proteoliposome swelling assay under improper conditions and its conclusion cannot be trusted (Nikaido & Vaara, 1985). It has been suggested that aminoglycosides, being polycations, perturb the polyanionic bilayer of OM, and diffuse across the OM in this manner (the self-promoted uptake) (Hancock, 1981). This pathway could indeed be significant in species containing porins of extremely low permeability, such as Pseudomonas aeruginosa (for an up-to-date view of the major porin of this species, see (Sugawara et al., 2010)).
Tetracyclines contain multiple charged groups, and are again unlikely to diffuse significantly through the OM bilayer. Simulations showed that they are likely to diffuse through enterobacterial porin channels as Mg2+-complexes (Thanassi et al., 1995)(Table 2). It seems likely that porins are also important for the OM diffusion of relatively small agents, such as fluoroquinolones and chloramphenicol (Table 2).
There are large, relatively lipophilic compounds that work poorly against gram-negative bacteria, for example macrolides, rifamycins, novobiocin, or fusidic acid. Their slow penetration across the enterobacterial OM must occur predominantly through the asymmetric lipid bilayer domain, as they are too large and too lipophilic to go through the porin channels (Table 2). This idea is experimentally confirmed by perturbing the LPS leaflet of the OM bilayer either by agents such as polymyxin B nonapeptide (Vaara, 1992)(Table 2) or genetically by mutations that affect the extreme proximal portion of the LPS structure (Vaara, 1993). These manipulations often produce huge (sometimes close to a thousandfold) decrease in the MIC values.
Given the importance of porins for the influx of agents that are effective (and thus useful) against gram-positive bacteria, it is unfortunate that we know so little about the overall permeability of porins in organisms other than E. coli and P. aeruginosa. We know, however, the permeation rates of small hydrophilic molecules through the porins of Enterobacter cloacae are significantly slower (by about fivefold) than through those of E. coli (Nikaido et al., 1990; Vu & Nikaido, 1985). In contrast, the porins of Enterobacter aerogenes, which is often treated by clinical microbiologists together with E. cloacae as Enterobacter spp., have high permeability similar to E. coli porins (Sugawara E, Nikaido H, and Pagès J-M, unpublished results). Perhaps this limited permeability of E. cloacae OM contributed to the predominance of constitutive ampC mutants of E. cloacae when the third-generation cephalosporins were first introduced. However, similar mutants of E. aerogenes were also isolated at that time, albeit in smaller numbers (Chow et al., 1991), showing that this relatively small differences in OM permeability may not produce absolute distinctions on the behavior of bacteria in the presence of antibiotics. In contrast, a large difference (of about two orders of magnitude) in OM permeability between P. aeruginosa and E. coli does produce a different behavior. For example, with the much lower OM permeability, efflux can be much more effective in P. aeruginosa. Thus the cephalosporin resistance patterns in many clinical isolates of P. aeruginosa could not be explained with the OM barrier and the periplasmic β-lactamase activity alone, and indeed led to the discovery of gram-negative multidrug efflux pumps that pump out β-lactams (Li et al., 1994). In contrast, the role of efflux pumps in β-lactam resistance in enterobacterial strains, with their highly permeable porins, has often escaped notice until a clever experimental setup was used very recently with K. pneumoniae (Pagès et al., 2009) (discussed above).
2. Decreased susceptibility due to down-regulation of porins
Resistance caused by the periplasmic β-lactamase or by the multidrug efflux pump(s) relies on the presence of OM barrier. Thus decreased expression of porins will often lead to decreased susceptibility. Already in 1981, selection of E. coli K-12 population with the dianionic compound carbenicillin was shown to result in mutants lacking the porin with a larger channel, OmpF (Harder et al., 1981)(Table 2). Dianionic drugs penetrate through porin channels slowly (see above), and thus the loss of OmpF produces a strong, 8- to 16-fold increase in their MIC. In contrast, the MIC of zwitterionic β-lactams (cephaloridine, ampicillin) remain essentially unaltered because they diffuse rapidly (see above) presumably even through the narrower OmpC channel.
This is not a situation found exclusively in the laboratory. Some years later, a ceftazidime-resistant, TEM-β-lactamase-producing E. coli strain was isolated from a patient receiving a monotherapy of ceftazidime, and was found to produce decreased levels of both OmpF and OmpC (Bakken et al., 1987). TEM-type β-lactamases, most commonly-encountered β-lactamases, are most often found in E. coli and K. pneumoniae, but they are also found in other species of Gram-negative bacteria with increasing frequency. Surprisingly the strain was not resistant to any other cephalosporins, but the detailed study of β-lactamase was not carried out. In the same year, a Salmonella typhimurium mutant lacking OmpC (and also producing a TEM-type enzyme) was selected in a patient during treatment with cephalexin (Medeiros et al., 1987). This particular observation at first seems to contradict the in vitro selection data in the preceding paragraph. Why should the deletion of smaller channel porin, OmpC, make a difference in MIC? Why should the use of cephalexin, a zwitterionic and rapidly penetrating cephalosporin, select a mutant of this type? The plausible explanation is that in a high osmolarity environment like human body, the expression of OmpF is repressed enough (see Nikaido & Vaara, 1985) so that only OmpC can function as a significant diffusion channel. Indeed, this OmpC-defective mutant selected in vivo were more resistant (4- to 8-fold higher MIC) to various cephalosporins, only when high osmolarity media (Mueller-Hinton with the osmolarity of 323 mM (Hanberger et al., 1990) or LB) were used. This situation emphasizes the fact that physiological effects of the environment on porin expression must be considered carefully.
In 1996, a survey of β-lactam-resistant enterobacterial isolates in France was made (Charrel et al., 1996), and showed that 44% of resistant strains of E. aerogenes had alterations in porin pattern, whereas only 6% had such changes in E. cloacae. Perhaps this difference is related to the lower permeability of porins in E. cloacae (discussed above), which might be enough to confer significant resistance to more recently developed cephalosporins that tend to be bulky and often contain two anionic groups. In contrast, E. aerogenes with its highly permeable porins needs their decreased expression for resistance. Although Omp35, an OmpF homolog presumably with a larger channel, is needed to keep E. aerogenes susceptible to ceftriaxone, a bulky dianionic drug, in low-osmolality media in the laboratory, the strains selected in the patients undergoing cephalosporin therapy usually had decreased level of OmpC homolog, Omp36 (Malléa et al., 1998), reminiscent of the observation with S. typhimurium mentioned above. It is difficult to pinpoint the antibiotics that led to the selection of these porin-deficient mutants in the clinical setting, but perhaps the agents that would overcome conventional resistance mechanisms, such as β-lactamase, could have been responsible. Possibly the widespread use of fourth-generation cephalosporins and the bulky carbapenems contributed to the selection of these mutants.
Interestingly, a discovery of mutated Omp36 in resistant clinical isolates (Chevalier et al., 1999), followed up by more detailed studies (De et al., 2001), established a novel mechanism for resistance. Thus a genetic modification of pore structure enabled the pathogens to resist antibiotic attack, while maintaining the adequate influx of nutrients, thus avoiding the physiological disadvantages brought about by the reduced levels of porins.
In the examples described so far, the OM permeation barrier acted synergistically with the β-lactamase-catalyzed hydrolysis of the drugs. However, for other drugs the second step could very well be the process of multidrug efflux, and the efflux produces very large increases in MIC especially when the drug influx across the OM is slow (see the strong sensitization to cloxacillin, novobiocin, and erythromycin upon deletion of acrAB genes (Table 2)). In a multidrug-resistant clinical isolate of K. pneumoniae overexpressing the AcrAB-TolC pump, a laboratory selection with ertapenem, a bulky carbapenem with two anionic and one cationic groups, resulted in the greatly decreased expression of the OmpC porin (Bialek et al., 2010). The porin-deficiency also increased cefoxitin MIC, suggesting the synergy between the OM barrier and the efflux pump activity. However, there was no increase in the MIC of ceftazidime and cefotaxime, an observation that is not easy to explain.
We also note that the major global regulator MarA, that makes a primary contribution to the increased expression of the main enterobacterial pump, AcrB, also represses the expression of the wider channel porin OmpF, further enhancing the synergy between the OM barrier and the efflux (Davin-Regli et al., 2008; Li & Nikaido, 2004).
In P. aeruginosa, the efflux pump activity is much more effective in increasing resistance levels, because its main porin, OprF, shows permeability about two orders of magnitude lower than E. coli OmpF (Sugawara et al., 2006; Yoshimura et al., 1983). This synergy is at the bottom of the much higher intrinsic resistance levels of P. aeruginosa for most classes of antibiotics, in comparison with enterobacterial strains. Thus permeabilization of the OM bilayer by treatment with PMBN results in a large decrease of MIC, for many drugs exceeding that caused by the inactivation of the main, constitutive multidrug efflux pump MexAB-OprM (Nikaido, 1998). Chloramphenicol MIC, for example, decreases fourfold upon inactivation of MexA whereas OM permeabilization decreases it eightfold. Tetracycline MIC decreases 32-fold and 16-fold, respectively, under the corresponding conditions. In contrast, for large and lipophilic compounds that must traverse the OM through its bilayer region, such as fusidic acid, novobiocin, and erythromycin, the permeabilization of this bilayer tends to have more spectacular results, whereas MexA inactivation has lesser effect, perhaps because the activity of other remaining efflux pumps is sufficient to pump out the small number of drug molecules that trickle through the wild-type OM.
The effectiveness of multidrug efflux in the presence of an effective OM barrier is also suggested by the important role the efflux plays in producing clinically relevant levels of resistance for many agents. For example, a survey on fluoroquinolone-resistant clinical isolates of P. aeruginosa from Japan found that in 96% of the strains MexAB pump overproduction played a major role in resistance (Cho et al., 1999). Also the first reports on the multidrug efflux pump(s) in this species (Li et al., 1994) were based on the study of clinical isolates, which suggest that such efflux-based resistance to a wide range of agents (including β-lactams) was already prevalent in the early 1990s. This forms a strong contrast to the situation in Enterobacteriaceae, where the contribution of active efflux to resistance was not often recognized, especially for β-lactams, until fairly recently (see, for example, Pagès et al., 2009).
3. Quantitative Analysis of Interaction between OM Barrier and Efflux: Situations in wild type E. coli K12
Our ultimate aim is to understand, in quantitative terms, how the interaction among OM barrier, periplasmic β-lactamase, and efflux affects the MIC values found in intact cells. E. coli K12 is the organism in which the major RND efflux pump AcrB (see above) as well as the barrier properties of its OM have been studied most intensively, and thus is the only system, currently, where an analysis of this type is possible.
(a) Penicillins and cephalosporins
Penicillins and cephalosporins contain a rather strong carboxylic acid group, and we believe that these compounds permeate across OM almost exclusively through porin channels (see above). As regards the diffusion of β-lactams through the general porin channels, it was found that at least some of these compounds interact specifically with a site within the porin channel (Danelon et al., 2006; James et al., 2009; Nestorovich et al., 2002; Pagès et al., 2008). However, whether this interaction is central to their permeation is still open to debate. In any case, MD simulation has shown the “binding” of ampicillin to the OmpF channel is extremely weak, with the association constant close to 1 M−1 (Hajjar et al., 2010). For comparison the association constant of maltopentaose to the maltooligosaccharide-specific channel LamB is 17,000 M−1 (Benz et al., 1987). For the present discussion, the importance of the “specific” permeation through the porin channel becomes significant only if this permeation pathway produces a saturation kinetics. However, in practice this should not be detectable, as we are dealing with the diffusion of drugs at less than a millimolar concentrations, in which the presence of very weak binding sites with the association constant of 1 M−1 should be totally invisible. We thus neglect the issue of the specific pathway in the discussion that follows, and treat the diffusion through non-specific porins as a spontaneous diffusion process following Fick’s first law of diffusion.
We have recently determined the efflux kinetics of cephalosporins (Nagano & Nikaido, 2009) and penicillins (Lim & Nikaido, 2010) in E. coli. Our approach has been already described above. However, in order to improve the precision of the assay, we had to “optimize” the system, introducing several conditions (larger porin channel and a slight overproduction of AcrAB) that do not exist in unmodified cells of E. coli K12. Under these conditions, the contribution of efflux was minimal at low external concentrations of cephalosporins that would mimic the clinical situations. For example, with cephamandole, at an external concentration of 22 μg mL−1, the number of substrate molecules hydrolyzed by the β-lactamase is three times higher than those removed by the efflux. This is largely due to the strong positive cooperativity of the efflux process, diminishing the pump contribution at lower substrate concentrations (Fig. 2).
We can estimate what happens in the wild type E. coli K-12. By using the kinetic constants of wild-type AmpC [in strain LA5 of (Nikaido & Normark, 1987)] and by assuming that the Vmax for efflux is about 1/3 of that in the marR strain used, we can calculate that at the periplasmic concentration of cefamandole that just inhibits cell growth [0.25 μg mL−1 or 0.54 μM] (Nikaido & Normark, 1987), the hydrolysis rate would be about 200-fold higher than the rate of efflux. A MIC values of cephalosporins rather accurately without considering the efflux process (Nikaido & Normark, 1987).
The situation becomes different with the penicillins (Lim & Nikaido, 2010). Here we used a strain modified in three areas. (a) In order to ensure rapid influx of the drugs, we used an E. coli mutant producing OmpC porin with an expanded channel (Misra & Benson, 1988). (b) In order to produce hydrolysis of various penicillins at a reasonable rate, oxa7 gene was introduced on a plasmid. (c) The synthesis of AcrB pump was enhanced [about three-fold (Ma et al., 1996)] by the inactivation of the acrR repressor gene. Thus although the kinetic parameters reported for efflux are believed to be correct (except the Vmax values that are inflated), other parameters such as penicillinase activity and porin permeability are very strongly distorted in our intentionally artificial construct.
We can calculate what happens to the MIC values of the wild-type K12 strain. For cloxacillin, because the strain lacking the AcrAB efflux pump as well as the OXA-7 penicillinase has an MIC of 4.9 3g mL−1 or roughly 10 3M, we assume that this is the periplasmic cloxacillin concentration that is needed to inhibit cell growth, i.e. the value of Cinh (Nikaido & Normark, 1987). Neglecting the hydrolysis by AmpC, which is in the undetectable range, the efflux rate Ve at this concentration (with the wild-type level expression of AcrAB) is calculated to be about 0.3 nmol mg−1 s−1 using the parameters listed (Lim & Nikaido, 2010). MIC of the wild-type strains are 512 (Nikaido et al., 1998) or 256 (Mazzariol et al., 2000) 3g mL−1. At 512 3g mL−1, the rate of inflow across OM is estimated as 0.3 nmol/mg/s, precisely balancing the efflux, if the permeability coefficient previously determined for benzylpenicillin [0.2 × 10−5 cm s−1] (Nikaido & Normark, 1987) is used. In contrast, for ampicillin, MIC of acrB mutant was 1 3g mL−1 (Lim & Nikaido, 2010) and that of ampC mutant 0.7 3g mL−1 (Nikaido & Normark, 1987). However, these mutants still contain the AmpC enzyme or the AcrB efflux, respectively, so that these values cannot be taken directly as Cinh. We can assume that the true Cinh is one-half of 0.7 3g mL−1, or 1 3M; this is close to the MIC of ampC acrB double mutant [0.5 3g mL−1] (Mazzariol et al., 2000). The efflux rate Ve at this periplasmic concentration is calculated to be 0.05 nmol mg−1 s−1. The rate of hydrolysis by the AmpC enzyme is also at the same level, based on the parameters reported earlier (Nikaido & Normark, 1987). At the external concentration of 2.5 3g mL−1, the rate of diffusion across the outer membrane is calculated as 0.08 nmol mg−1 s−1, using the previously determined value of permeability coefficient [9.8 × 10−5 cm s−1] (Nikaido & Normark, 1987), and it is close to the sum of hydrolysis and efflux. The MIC of the wild-type K12 was reported to 2.5 (Okusu et al., 1996) or 2 (Mazzariol et al., 2000) 3g mL−1, close to the value obtained by the above theoretical prediction.
This analysis gives us important insights. Since the discovery of their multidrug efflux function more than a decade ago, kinetic parameters of RND-pump-mediated drug efflux have never been elucidated until these studies of cephalosporin and penicillin efflux (Lim & Nikaido, 2010; Nagano & Nikaido, 2009). Thus the only measure for the efficiency of efflux of any drug was to compare the MIC of the wild type strain with that of the pump-deficient mutant. With this criterion, isoxazolylpenicillins such as cloxacillin appear to be a very strong substrate of the AcrB pump, because the genetic deletion of the pump decreases MIC 256-fold or more (Mazzariol et al., 2000; Nikaido et al., 1998). In contrast, compounds like ampicillin were always thought to be very poor substrates of the pump, because AcrB deletion lowers MIC only twofold. However, our study of efflux kinetics (Lim & Nikaido, 2010) showed that both cloxacillin and ampicillin are similarly good substrates of the pump: the K0.5 values are identical, and there is only about twofold difference in Vmax. Thus it is clear that the ratios of MICs do not serve even as a crude index of the efflux efficiency of any given drug. So if ampicillin and cloxacillin are both pumped out well by AcrB, why does deletion of the pump gene produce such a spectacular change in MIC only with cloxacillin? Scrutiny of the calculations given above shows that the major factor is the poor permeability of the lipophilic isoxazolylpenicillins (although an additional factor is the contribution of AmpC-catalyzed hydrolysis of ampicillin, but not cloxacillin). Thus, as was proposed earlier (Nikaido, 2001), what makes RND multidrug efflux pumps so effective is the synergy with the OM barrier. This point is also confirmed by the observation that with the large channel mutant porin, the cloxacillin MIC of the strain containing the plasmid-coded oxa7 gene goes down only 2.5-fold upon deletion of the acrAB genes (Lim & Nikaido, 2010).
(b) Tetracyclines
So far, β-lactams are the only compounds whose periplasmic concentrations Cp can be determined experimentally. However, with tetracycline we attempted a rather careful simulation of its entry and efflux with several experimentally determined parameters (Thanassi et al., 1995). When this study was done in 1995, the identity of the major constitutive efflux pump was still uncertain, and thus acrAB deletion mutants were not used. Nevertheless, the model illustrates what may happen in the interaction of tetracycline with the wild-type E. coli cells. We first built a model for a porin-deficient E. coli strain B/r. Since only 3.7% of the tetracycline molecule in the medium was calculated to be present in the uncharged form, and since the permeability of the asymmetric OM bilayer was earlier found to be about 1% of that of the conventional phospholipid bilayer (Plesiat & Nikaido, 1992), we could estimate the permeability coefficient of tetracycline across the porinless OM as about 1 × 10−7 cm s−1. Tetracycline then diffuses rapidly into the cytosol. It is also pumped out by a constitutive pump, now known to be AcrAB. We do not know the Km of AcrAB for tetracycline, but since McMurry et al. (McMurry et al., 1982) found the Km of the endogenous efflux system (i.e. AcrAB) to be above 100 μM, we assumed 200 μM. Thus in this simulation, the only adjustable parameter was the Vmax of efflux mediated by AcrAB. The two simultaneous differential equations related to the time-dependent change of the periplasmic drug concentration, Cp, and the cytosolic drug concentration, Cc, were solved for various values of Vmax, and 0.003 nmol/mg/s gave a good fit to the experimentally observed time course of tetracycline accumulation. Compared with the experimentally determined Vmax values for the efflux of β-lactams, usually in the range of 0.3–1 nmol mg−1 s−1 (Lim & Nikaido, 2010; Nagano & Nikaido, 2009), this value seems too low. One likely reason for this discrepancy is our low estimate of the permeability of the OM bilayer. We assumed that the OM bilayer has a permeability coefficient of only 1% of that of the conventional phospholipid bilayer according to our old data (Plesiat & Nikaido, 1992), but this estimate is now known to be too low because the probes used, sterols, were found to be pumped out by the endogenous AcrAB pump (Elkins & Mullis, 2006).
Another factor in this simulation is that the endogenous pump (AcrAB) was supposed to capture tetracycline from within the cytoplasm, whereas we now believe that the substrate capture is likely to occur exclusively from the periplasm (Nikaido & Takatsuka, 2009). Tetracycline is pumped out into the periplasm by endogenous major facilitator superfamily (MFS) pumps such as MdfA (Fluman & Bibi, 2009), and then captured in periplasm by AcrAB, as was shown first by Lomovskaya’s group (Lee et al., 2000) and also later by Schuldiner’s group (Tal & Schuldiner, 2009). Our use of the incorrect model thus could have contributed to the inaccuracy of prediction.
Sigler et al. devised an elegant assay for permeation of tetracycline by using its fluorescence enhancement caused by binding to the TetR repressor, and came up with the permeability coefficient of 2.4 × 10−9 cm s−1 for its permeation across phospholipid bilayer (Sigler et al., 2000. This value is several orders of magnitude lower than the permeability coefficient of IM we calculated from its octanol/water partition coefficient and size, around 1 × 10−5 cm/s (Thanassi et al., 1995). Furthermore, the permeability coefficient of Sigler et al. predicts half-equilibration time across IM of 35 min, yet in our hands the half-equilibration was achieved within 1 or 2 min even in intact cells with the additional OM barrier (Thanassi et al., 1995). The equilibration is even faster in the experiments of McMurry et al. (McMurry et al., 1982). It is unclear how the experiments of Sigler et al. led to such low estimates that are inconsistent with observations in other laboratories, but tetracycline is fluorescent in a free state, and their experiments could have involved very high background fluorescence that may have compromised the accuracy of their assay.
(c) Macrolides
Mutations in the target of macrolides, ribosomes, often lower their affinity to the drug. Ehrenberg’s laboratory reported that such mutants showed increased resistance in the efflux-proficient background, but in the pump-deficient strains its MIC level remained at a similar low level as that of the strain with unaltered ribosomes (Lovmar et al., 2009). In an effort to understand this “target masking” phenomenon by the absence of efflux pump, a series of simulations were made (Fange et al., 2009; Lovmar et al., 2009), which finally led to the conclusion that the presence of two stable states (“bi-stability”) is the cause of these phenomena.
The conclusion is interesting, but for this author (H. N.) is not yet completely persuasive. First, the simulation was done with too many adjustable parameters with arbitrary values (permeability across OM, permeability across IM, the Km and Vmax of the efflux pump) with none of them experimentally determined. Under such conditions, any model can be made to fit the sole set of the observations, i.e. the erythromycin susceptibility of the whole E. coli cells. Second, some of the assumed parameters are quite unrealistic. For example, from the logP value (2.54) and the weakness of its basic group [pKa 8.6] (Martin et al., 1972), the permeability coefficient of erythromycin across IM is expected to be about 1 × 10−3 cm s−1 following the procedure used by us (Thanassi et al., 1995). Yet the parameter used [0.1 s−1 as the first-order rate constant] (Lovmar et al., 2009) corresponds to the permeability coefficient of 2.3 × 10−6 cm s−1, nearly three orders of magnitude lower than the expected value above. Third, scrutiny of the growth inhibition data tends to put the “target masking” phenomenon itself on a shaky ground. Thus in the data of Lovmar et al. (Lovmar et al., 2009) erythromycin concentrations giving 50% growth inhibition increased from 100 to 500 μM upon the introduction of the ribosomal mutation L22 (Δ82–84) in the wild-type background. Although the difference became superficially less impressive in the ΔacrB mutant, the approximately fivefold ratio of the 50% inhibitory concentrations [3.5 vs. 20 μM] was unchanged. The ratio may have decreased to about two in ΔtolC mutants, but here not enough points were taken and ΔtolC mutants are known to have unexplained growth defects even in the absence of antibiotics (Dhamdhere & Zgurskaya, 2010).
V. Conclusion
Dissecting the molecular and conformational steps of the drug transport through the bacterial efflux pump is an exciting challenge. Regarding the mechanistic and struture-activity relathionship aspects of the process and with the continuing clinical and health impact of MDR phenotype, this represents an important direction taking into account the scarcity of new antibiotics in the pipeline. This is especially the case with Gram-negative bacteria that exhibit a sophisticated envelope and a complex architecture of membrane transporters.
The clinical investigations found that the AcrAB-MexAB family is the main type of drug efflux pump reported in human infectious diseases. A large number of enterobacterial clinical isolates collected during the antibiotic therapy of infected/colonized patients exhibit a significant overproduction of the pump type. Consequently, it is of interest to elucidate the functional steps of the pump and the affinity for substrates of interest. At the moment, a variety of compounds (natural or chemically produced) have been shown to be recognized and translocated by AcrAB pumps. The polyselectivity of AcrAB pump group in clinical isolates is the basis of the low susceptibility towards a large collection of antibacterial agents (Li & Nikaido, 2009; Poole, 2007; Piddock, 2006).
Crystallographic studies on the asymmetric AcrB and MexB trimers identified the substrate-binding pocket deep in the periplasmic domain, which presumably plays a decisive role in determining the specificity of the pump. Computational and biochemical studies revealed that different antimicrobials and EPIs bind to different sub-domains within this large pocket, and that only those ligands binding to the same sub-domain appeared to compete with each other for efflux. However, the roles of subsidiary binding sites (other than the deep pocket mentioned above) and the actual translocation process for drugs need more studies. For example, although EPIs and drugs both bind to the pocket well, why do only EPIs produce a strong inhibition of efflux?
Examination of the kinetics of β-lactam efflux through AcrB, especially in the context of intact E. coli cells, produced significant insights. Thus although cloxacillin and ampicillin are pumped out with similar kinetic constants, the efflux reduces the susceptibility of E. coli very strongly only with cloxacillin, mainly because it diffuses through the porin channel much more slowly than ampicillin. We must therefore consider OM permeability always together with active efflux in order to understand the resistance of gram-negative bacteria.
With our knowledge of both the significance of efflux in clinical isolates and the molecular details of pump-ligand interaction, we can consider the steps to combat the now widespread, broad substrate-range, efflux process.
The first possibility is to improve the design of the antibiotic molecule in order to reduce its susceptibility to efflux. With the extremely flexible structure of the binding site, this may at first appear to be just a daydream. However, the knowledge that simply exchanging the penicillin nucleus with the cephalosporin nucleus drastically decreases the apparent affinity of drugs to AcrB (Lim et al., 2010; Nagano & Nikaido, 2009) suggests that this approach should not be abandoned so easily. Of course even with cephalosporins the slow permeation through the porin channel would make efflux significant, and we desperately need more knowledge on this permeation process (see IV.3.a).
The second approach is the development of EPI. Although EPIs in the past have been discovered and refined by screening efforts, now that we know the structure of the critical binding pocket we may be able to utilize our knowledge on their detailed interaction with the transporter. Furthermore, our knowledge on the specificity of competition between EPI and the drug substrate (Pagès et al., 2009; Takatsuka et al., 2010) may lead to the development of perhaps more effective EPIs of somewhat more limited spectrum. These new agents may be able to rejuvenate the activity of “old antibiotics” against MDR bacteria. We note that this kind of combination therapy has already been quite successful in other areas, in inhibiting the enzymatic degradation of β-lactams by clavulanate or tazobactam.
Nowadays, it is clear that affinity, kinetic and dynamic parameters are key elements for the rational development of new antibacterial agents capable to circumvent the membrane barrier strategy which controls the drug influx and efflux in Gram-negative bacteria.
Acknowledgments
We gratefully thank Anne Favard for the preparation of this manuscript. This study was supported in Marseille by the Service de Santé des Armées, by the Université de la Méditerranée and by ANR METABACT, and in Berkeley by a USPHS grant AI-09644. We apologize to those whose papers and studies could not be cited owing to space limitations.
References
- Aathithan S, French GL. Prevalence and role of efflux pump activity in ciprofloxacin resistance in clinical isolates of Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2011 Feb 1; doi: 10.1007/s10096-010-1147-0. [DOI] [PubMed] [Google Scholar]
- Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Amábile-Cuevas CF, Arredondo-García JL, Cruz A, Rosas I. Fluoroquinolone resistance in clinical and environmental isolates of Escherichia coli in Mexico City. J Appl Microbiol. 2010;108(1):158–162. doi: 10.1111/j.1365-2672.2009.04401.x. [DOI] [PubMed] [Google Scholar]
- Amaral L, Pagès JM, Fanning S. Efflux pumps of Gram-negative bacteria: genetic responses to stress and the modulation of their activity by pH, inhibitors and phenothaizines. Advances In Enzymology and Related Areas of Molecular Biology. 2011;77:61–108. doi: 10.1002/9780470920541.ch2. [DOI] [PubMed] [Google Scholar]
- Amicosante G, Oratore A, Franceschini N, Maccarrone M, Strom R, Galleni M, Frere JM. Citrobacter diversus ULA-27 β-lactamases. Improved purification and general properties. Biochem J. 1988;254:885–890. doi: 10.1042/bj2540885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken JS, Sanders CC, Thomson KS. Selective ceftazidime resistance in Escherichia coli: Association with changes in outer membrane protein. J Infect Dis. 1987;155:1220–1225. doi: 10.1093/infdis/155.6.1220. [DOI] [PubMed] [Google Scholar]
- Bal AM, Kumar A, Gould IM. Antibiotic heterogeneity: from concept to practice. Ann N Y Acad Sci. 2010;1213:81–91. doi: 10.1111/j.1749-6632.2010.05867.x. [DOI] [PubMed] [Google Scholar]
- Baum EZ, Crespo-Carbone SM, Morrow BJ, Davies TA, Foleno BD, He W, Queenan AM, Bush K. Effect of MexXY overexpression on ceftobiprole susceptibility in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(7):2785–2790. doi: 10.1128/AAC.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin T, Aaron SD, Giesbrecht-Lewis T, Vandemheen K, Mah TF. Characterization of clonal strains of Pseudomonas aeruginosa isolated from cystic fibrosis patients in Ontario, Canada. Can J Microbiol. 2010;56:548–557. doi: 10.1139/w10-043. [DOI] [PubMed] [Google Scholar]
- Becnel Boyd L, Maynard MJ, Morgan-Linnell SK, Horton LB, Sucgang R, Hamill RJ, Jimenez JR, Versalovic J, Steffen D, Zechiedrich L. Relationships among ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin MICs for fluoroquinolone-resistant Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009;53:229–234. doi: 10.1128/AAC.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz R, Schmid A, Vos-Scheperkeuter GH. Mechanism of sugar transport through the sugar-specific LamB channel of Escherichia coli outer membrane. J Membr Biol. 1987;100:21–29. doi: 10.1007/BF02209137. [DOI] [PubMed] [Google Scholar]
- Bialek S, Lavigne JP, Chevalier J, Marcon E, Leflon-Guibout V, Davin A, Moreau R, Pagès JM, Nicolas-Chanoine MH. Membrane efflux and influx modulate both multidrug resistance and virulence of Klebsiella pneumoniae in a Caenorhabditis elegans model. Antimicrob Agents Chemother. 2010;54:4373–4378. doi: 10.1128/AAC.01607-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek S, Lavigne JP, Chevalier J, Marcon E, Leflon-Guibout V, Davin A, Moreau R, Pagès JM, Nicolas-Chanoine MH. Membrane efflux and influx modulate both multidrug resistance and virulence of Klebsiella pneumoniae in a Caenorhabditis elegans model. Antimicrob Agents Chemother. 2010;54:4373–4378. doi: 10.1128/AAC.01607-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blot S, Depuydt P, Vandewoude K, De Bacquer D. Measuring the impact of multidrug resistance in nosocomial infection. Curr Opin Infect Dis. 2007;20:391–396. doi: 10.1097/QCO.0b013e32818be6f7. [DOI] [PubMed] [Google Scholar]
- Bohnert JA, Kern WV. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother. 2005;49:849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert JA, Schuster S, Seeger MA, Fahnrich E, Pos KM, Kern WV. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol. 2008;190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornet C, Davin-Regli A, Bosi C, Pagès JM, Bollet C. Imipenem resistance of Enterobacter aerogenes mediated by outer membrane permeability. J Clin Microbiol. 2000;38:1048–1052. doi: 10.1128/jcm.38.3.1048-1052.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornet C, Chollet R, Malléa M, Chevalier J, Davin-Regli A, Pagès JM, Bollet C. Imipenem and expression of multidrug efflux pump in Enterobacter aerogenes. Biochem Biophys Res Commun. 2003;301:985–990. doi: 10.1016/s0006-291x(03)00074-3. [DOI] [PubMed] [Google Scholar]
- Boutoille D, Corvec S, Caroff N, Giraudeau C, Espaze E, Caillon J, Plésiat P, Raynaud A. Detection of an IS21 insertion sequence in the mexR gene of Pseudomonas aeruginosa increasing β-lactam resistance. FEMS Microbiol Lett. 2004;230:143–146. doi: 10.1016/S0378-1097(03)00882-6. [DOI] [PubMed] [Google Scholar]
- Bratu S, Landman D, George A, Salvani J, Quale J. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother. 2009;64:278–283. doi: 10.1093/jac/dkp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel RN, Pagès JM, De Micco P, Mallea M. Prevalence of outer membrane porin alteration in β-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother. 1996;40:2854–2858. doi: 10.1128/aac.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau SL, Chu YW, Houang ET. Novel resistance-nodulation-cell division efflux system AdeDE in Acinetobacter genomic DNA group 3. Antimicrob Agents Chemother. 2004;48:4054–4055. doi: 10.1128/AAC.48.10.4054-4055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier J, Pagès JM, Mallea M. In vivo modification of porin activity conferring antibiotic resistance to Enterobacter aerogenes. Biochem Biophys Res Comm. 1999;266:248–251. doi: 10.1006/bbrc.1999.1795. [DOI] [PubMed] [Google Scholar]
- Chevalier J, Bredin J, Mahamoud A, Malléa M, Barbe J, Pagès JM. Inhibitors of antibiotic efflux in resistant Enterobacter aerogenes and Klebsiella pneumoniae strains. Antimicrob Agents Chemother. 2004;48:1043–1046. doi: 10.1128/AAC.48.3.1043-1046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier J, Mulfinger C, Garnotel E, Nicolas P, Davin-Régli A, Pagĕs JM. Identification and evolution of drug efflux pump in clinical Enterobacter aerogenes strains isolated in 1995 and 2003. PLoS ONE. 2008;3:e3203. doi: 10.1371/journal.pone.0003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier J, Mahamoud A, Baitiche M, Adam E, Viveiros M, Smarandache A, Militaru A, Pascu ML, Amaral L, Pagès JM. Quinazoline derivatives are efficient chemosensitizers of antibiotic activity in Enterobacter aerogenes, Klebsiella pneumoniae and Pseudomonas aeruginosa resistant strains. Int J Antimicrob Agents. 2010;36:164–168. doi: 10.1016/j.ijantimicag.2010.03.027. [DOI] [PubMed] [Google Scholar]
- Chiu CH, Lee HY, Tseng LY, Chen CL, Chia JH, Su LH, Liu SY. Mechanisms of resistance to ciprofloxacin, ampicillin/sulbactam and imipenem in Acinetobacter baumannii clinical isolates in Taiwan. Int J Antimicrob Agents. 2010;35:382–386. doi: 10.1016/j.ijantimicag.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Cho D, Blais J, Tangen K, Ford C, Lee A, Lomovskaya O, Chamberland S, Miller GH. Prevalence of efflux pumps among clinical isolates of fluoroquinolone-resistant Pseudomonas aeruginosa. Abstracts, 39th Interscience Conference on Antimicrobial Agents and Chemotherapy; Sep 26–29; 1999. p. 327. (abstract n°1267) [Google Scholar]
- Chopra I, Schofield C, Everett M, O’Neill A, Miller K, Wilcox M, Frère JM, Dawson JM, Czaplewski L, Urleb U, Courvalin P. Treatment of health-care-associated infections caused by Gram-negative bacteria: a consensus statement. Lancet Infect Dis. 2008;8:133–139. doi: 10.1016/S1473-3099(08)70018-5. [DOI] [PubMed] [Google Scholar]
- Chow JW, Fine MJ, Shlaes DM, Quinn JP, Hooper DC, Johnson MP, Ramphal R, Wagener MM, Miyashiro DK, Yu VL. Enterobacter bacteremia: Clinical features and emergence of antibiotic resistance during therapy. Ann Int Med. 1991;115:585–590. doi: 10.7326/0003-4819-115-8-585. [DOI] [PubMed] [Google Scholar]
- Chu YW, Chau SL, Houang ET. Presence of active efflux systems AdeABC, AdeDE and AdeXYZ in different Acinetobacter genomic DNA groups. J Med Microbiol. 2006;55(Pt 4):477–478. doi: 10.1099/jmm.0.46433-0. [DOI] [PubMed] [Google Scholar]
- Coyne S, Courvalin P, Périchon B. Efflux-Mediated Antibiotic Resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2010;55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damier-Piolle L, Magnet S, Brémont S, Lambert T, Courvalin P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumanii. Antimicrob Agents Chemother. 2008;52:557–562. doi: 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danelon C, Nestorovich EM, Winterhalter M, Ceccarelli M, Bezrukov SM. Interaction of zwitterionic penicillins with the OmpF channel facilitates their translocation. Biophys J. 2006;90:1617–1627. doi: 10.1529/biophysj.105.075192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin-Regli A, Bolla JM, James CE, Lavigne JP, Chevalier J, Garnotel E, Molitor A, Pagès JM. Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr Drug Targets. 2008;9:750–759. doi: 10.2174/138945008785747824. [DOI] [PubMed] [Google Scholar]
- Davin-Regli A, Bolla JM, James CE, Lavigne JP, Chevalier J, Garnotel E, Molitor A, Pagès JM. Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr Drug Targets. 2008;9:750–759. doi: 10.2174/138945008785747824. [DOI] [PubMed] [Google Scholar]
- Dé E, Basle A, Jaquinod M, Saint N, Mallea M, Molle G, Pagès JM. A new mechanism of antibiotic resistance in enterobacteriaceae induced by a structural modification of the major porin. Mol Microbiol. 2001;41:189–198. doi: 10.1046/j.1365-2958.2001.02501.x. [DOI] [PubMed] [Google Scholar]
- Dhamdhere G, Zgurskaya HI. Metabolic shutdown in Escherichia coli cells lacking the outer membrane channel TolC. Mol Microbiol. 2010;77:743–754. doi: 10.1111/j.1365-2958.2010.07245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D, Klepsch MM, Newstead S, Flaig R, De Gier JW, Iwata S, Beis K. The structure of the efflux pump AcrB in complex with bile acid. Mol Membr Biol. 2008;25:677–682. doi: 10.1080/09687680802552257. [DOI] [PubMed] [Google Scholar]
- Dunham SA, McPherson CJ, Miller AA. The relative contribution of efflux and target gene mutations to fluoroquinolone resistance in recent clinical isolates of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 2010;29(3):279–288. doi: 10.1007/s10096-009-0852-z. [DOI] [PubMed] [Google Scholar]
- Eicher T, Brandstätter L, Pos KM. Structural and functional aspects of the multidrug efflux pump AcrB. Biol Chem. 2009 Aug;390(8):693–9. doi: 10.1515/BC.2009.090. [DOI] [PubMed] [Google Scholar]
- Elkins CA, Mullis LB. Mammalian steroid hormones are substrates for the major RND-and MFS-type tripartite multidrug efflux pumps of Escherichia coli. J Bacteriol. 2006;188:1191–1195. doi: 10.1128/JB.188.3.1191-1195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins CA, Nikaido H. Substrate specificity of the rnd-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J Bacteriol. 2002;184:6490–6498. doi: 10.1128/JB.184.23.6490-6498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas ME, Bliziotis IA. Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int J Antimicrob Agents. 2007;29:630–636. doi: 10.1016/j.ijantimicag.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Fange D, Nilsson K, Tenson T, Ehrenberg M. Drug efflux pump deficiency and drug target resistance masking in growing bacteria. Proc Natl Acad Sci U S A. 2009;106:8215–8220. doi: 10.1073/pnas.0811514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L, Breidenstein EB, Hancock RE. Creeping baselines and adaptive resistance to antibiotics. Drug Resist Updat. 2011;14:1–21. doi: 10.1016/j.drup.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbain J, Peleg AY. Treatment of Acinetobacter infections. Clin Infect Dis. 2010;51:79–84. doi: 10.1086/653120. [DOI] [PubMed] [Google Scholar]
- Fluman N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim Biophys Acta. 2009;1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- Gandhi TN, DePestel DD, Collins CD, Nagel J, Washer LL. Managing antimicrobial resistance in intensive care units. Crit Care Med. 2010;38(8 Suppl):S315–23. doi: 10.1097/CCM.0b013e3181e6a2a4. [DOI] [PubMed] [Google Scholar]
- Ghisalberti D, Mahamoud A, Chevalier J, Baitiche M, Martino M, Pagès JM, Barbe J. Chloroquinolines block antibiotic efflux pumps in antibiotic-resistant Enterobacter aerogenes isolates. Int J Antimicrob Agents. 2006;27:565–569. doi: 10.1016/j.ijantimicag.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Giamarellou H, Poulakou G. Multidrug-resistant Gram-negative infections: what are the treatment options? Drugs. 2009;69:1879–1901. doi: 10.2165/11315690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Glatz K, Tóth A, Pászti J. The cyclohexane tolerance and Phe-Arg-β-naphtylamide susceptibility of multidrug-resistant Enterobacter cloacae clinical isolates, and the predominance of one PFGE clone in Hungary. Clin Microbiol Infect. 2010 doi: 10.1111/j.1469-0691.2010.03408.x. [DOI] [PubMed] [Google Scholar]
- Gwynn MN, Portnoy A, Rittenhouse SF, Payne DJ. Challenges of antibacterial discovery revisited. Ann N Y Acad Sci. 2010;1213:5–19. doi: 10.1111/j.1749-6632.2010.05828.x. [DOI] [PubMed] [Google Scholar]
- Hajjar E, Bessonov A, Molitor A, Kumar A, Mahendran KR, Winterhalter M, Pagès JM, Ruggerone P, Ceccarelli M. Toward screening for antibiotics with enhanced permeation properties through bacterial porins. Biochemistry. 2010;49:6928–6935. doi: 10.1021/bi100845x. [DOI] [PubMed] [Google Scholar]
- Hanberger H, Nilsson LE, Kihlstrom E, Maller R. Postantibiotic effect of β-lactam antibiotics on Escherichia coli evaluated by bioluminescence assay of bacterial ATP. Antimicrob Agents Chemother. 1990;34:102–106. doi: 10.1128/aac.34.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE. Aminoglycoside uptake and mode of action-with special reference to streptomycin and gentamicin. Il. Effects of aminoglycosides on cells. J Antimicrob Chemother. 1981;8:429–445. doi: 10.1093/jac/8.6.429. [DOI] [PubMed] [Google Scholar]
- Harder KJ, Nikaido H, Matsuhashi M. Mutants of Escherichia coli that are resistant to certain β-lactam compounds lack the OmpF porin. Antimicrob Agents Chemother. 1981;20:549–552. doi: 10.1128/aac.20.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasdemir UO, Chevalier J, Nordmann P, Pagès JM. Detection and prevalence of active drug efflux mechanism in various multidrug-resistant Klebsiella pneumoniae strains from Turkey. J Clin Microbiol. 2004;42:2701–2706. doi: 10.1128/JCM.42.6.2701-2706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegstad K, Langsrud S, Lunestad BT, Scheie AA, Sunde M, Yazdankhah SP. Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health? Microb Drug Resist. 16:91–104. doi: 10.1089/mdr.2009.0120. [DOI] [PubMed] [Google Scholar]
- Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10:441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquet D, Muller A, Blanc K, Plésiat P, Talon D, Monnet DL, Bertrand X. Relationship between antibiotic use and incidence of MexXY-OprM overproducers among clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52:1173–1175. doi: 10.1128/AAC.01212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquet D, Nordmann P, El Garch F, Cabanne L, Plésiat P. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2006;50:1347–1351. doi: 10.1128/AAC.50.4.1347-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquet D, Roussel-Delvallez M, Cavallo JD, Plésiat P. MexAB-OprM- and MexXY-overproducing mutants are very prevalent among clinical strains of Pseudomonas aeruginosa with reduced susceptibility to ticarcillin. Antimicrob Agents Chemother. 2007;51:1582–1583. doi: 10.1128/AAC.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsey M, Ellington MJ, Doumith M, Scott G, Livermore DM, Woodford N. Emergence of AcrAB-mediated tigecycline resistance in a clinical isolate of Enterobacter cloacae during ciprofloxacin treatment. Int J Antimicrob Agents. 2010;35:478–481. doi: 10.1016/j.ijantimicag.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Houang ET, Chu YW, Chu KY, Ng KC, Leung CM, Cheng AF. Significance of genomic DNA group delineation in comparative studies of antimicrobial susceptibility of Acinetobacter spp. Antimicrob Agents Chemother. 2003;47:1472–1475. doi: 10.1128/AAC.47.4.1472-1475.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain F, Nikaido H. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol Microbiol. 2010;78:320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Oh H, Jalal S, Karpati F, Ciofu O, Høiby N, Wretlind B. Chromosomal mechanisms of aminoglycoside resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clin Microbiol Infect. 2009;15:60–66. doi: 10.1111/j.1469-0691.2008.02097.x. [DOI] [PubMed] [Google Scholar]
- James CE, Mahendran KR, Molitor A, Bolla JM, Bessonov AN, Winterhalter M, Pagès JM. How β-lactam antibiotics enter bacteria: A dialogue with the porins. PLoS One. 2009;4:e5453. doi: 10.1371/journal.pone.0005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. News and Analysis: The antibacterial lead discovery challenge. Nature Reviews Drug Discovery. 2010;9:751–752. doi: 10.1038/nrd3289. [DOI] [PubMed] [Google Scholar]
- Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis. 2010;51(Suppl 1):S81–87. doi: 10.1086/653053. [DOI] [PubMed] [Google Scholar]
- Källman O, Giske CG, Samuelsen Ø, Wretlind B, Kalin M, Olsson-Liljequist B. Interplay of efflux, impermeability, and AmpC activity contributes to cefuroxime resistance in clinical, non-ESBL-producing isolates of Escherichia coli. Microb Drug Resist. 2009;15:91–95. doi: 10.1089/mdr.2009.0883. [DOI] [PubMed] [Google Scholar]
- Källman O, Motakefi A, Wretlind B, Kalin M, Olsson-Liljequist B, Giske CG. Cefuroxime non-susceptibility in multidrug-resistant Klebsiella pneumoniae overexpressing ramA and acrA and expressing ompK35 at reduced levels. J Antimicrob Chemother. 2008;62:986–990. doi: 10.1093/jac/dkn296. [DOI] [PubMed] [Google Scholar]
- Kern WV, Steinke P, Schumacher A, Schuster S, von Baum H, Bohnert JA. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J Antimicrob Chemother. 2006;57:339–343. doi: 10.1093/jac/dki445. [DOI] [PubMed] [Google Scholar]
- Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect. 2009;73:338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim SH, Jeon SM, Park MS, Rhie HG, Lee BK. Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clin Microbiol Infect. 2008;14:760–765. doi: 10.1111/j.1469-0691.2008.02033.x. [DOI] [PubMed] [Google Scholar]
- Kiser TH, Obritsch MD, Jung R, MacLaren R, Fish DN. Efflux pump contribution to multidrug resistance in clinical isolates of Pseudomonas aeruginosa. Pharmacotherapy. 2010;30:632–638. doi: 10.1592/phco.30.7.632. [DOI] [PubMed] [Google Scholar]
- Komp Lindgren P, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother. 2003;47:3222–3232. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreusch A, Neubuser A, Schiltz E, Weckesser J, Schulz GE. Structure of the membrane channel porin from Rhodopseudomonas blastica at 2.0 Å resolution. Prot Sci. 1994;3:58–63. doi: 10.1002/pro.5560030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman D, Bratu S, Quale J. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol. 2009;58(Pt 10):1303–1308. doi: 10.1099/jmm.0.012575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenbach E, Fishman NO, Metlay JP, Mao X, Bilker WB, Tolomeo P, Nachamkin I. Phenotypic and genotypic characterization of fecal Escherichia coli isolates with decreased susceptibility to fluoroquinolones: results from a large hospital-based surveillance initiative. J Infect Dis. 2006;194:79–85. doi: 10.1086/503046. [DOI] [PubMed] [Google Scholar]
- Lautenbach E, Metlay JP, Mao X, Han X, Fishman NO, Bilker WB, Tolomeo P, Wheeler M, Nachamkin I. The prevalence of fluoroquinolone resistance mechanisms in colonizing Escherichia coli isolates recovered from hospitalized patients. Clin Infect Dis. 2010;51:280–285. doi: 10.1086/653931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Mao W, Warren MS, Mistry A, Hoshino K, Okamura R, Ishida H, Lomovskaya O. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol. 2000;182:3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Mao W, Warren MS, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol. 2000;182:3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SB. Active efflux, a common mechanism for biocide and antibiotic resistance. Symp Ser Soc Appl Microbiol. 2002;31:65S–71S. [PubMed] [Google Scholar]
- Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Livermore DM, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: Resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother. 1994;38:1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SP, Nikaido H. Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob Agents Chemother. 2010;54:1800–1806. doi: 10.1128/AAC.01714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic--a vision for applied use. Biochem Pharmacol. 2006;71:910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovmar M, Nilsson K, Lukk E, Vimberg V, Tenson T, Ehrenberg M. Erythromycin resistance by L4/L22 mutations and resistance masking by drug efflux pump deficiency. Embo J. 2009;28:736–744. doi: 10.1038/emboj.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn AD, Fàbrega A, Sánchez-Céspedes J, Vila J. Prevalence of mechanisms decreasing quinolone-susceptibility among Salmonella spp. clinical isolates. Int Microbiol. 2010;13:15–20. doi: 10.2436/20.1501.01.107. [DOI] [PubMed] [Google Scholar]
- Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. The local repressor AcrR plays a modulating role in the regulation of AcrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamoud A, Chevalier J, Baitiche M, Adam E, Pagès JM. An alkylaminoquinazoline restores antibiotic activity in Gram-negative resistant isolates. Microbiology. 2011;157(Pt 2):566–571. doi: 10.1099/mic.0.045716-0. [DOI] [PubMed] [Google Scholar]
- Malléa M, Chevalier J, Bornet C, Eyraud A, Davin-Regli A, Bollet C, Pagès JM. Porin alteration and active efflux: Two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology. 1998;144 (Pt 11):3003–3009. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- Malléa M, Mahamoud A, Chevalier J, Alibert-Franco S, Brouant P, Barbe J, Pagès JM. Alkylaminoquinolines inhibit the bacterial antibiotic efflux pump in multidrug-resistant clinical isolates. Biochem J. 2003;376(Pt 3):801–805. doi: 10.1042/BJ20030963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamelli L, Prouzet-Mauléon V, Pagès JM, Mégraud F, Bolla JM. Molecular basis of macrolide resistance in Campylobacter: role of efflux pumps and target mutations. J Antimicrob Chemother. 2005;56:491–497. doi: 10.1093/jac/dki253. [DOI] [PubMed] [Google Scholar]
- Mao W, Warren MS, Black DS, Satou T, Murata T, Nishino T, Gotoh N, Lomovskaya O. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: The large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol Microbiol. 2002;46:889–901. doi: 10.1046/j.1365-2958.2002.03223.x. [DOI] [PubMed] [Google Scholar]
- Maragakis LL. Recognition and prevention of multidrug-resistant Gram-negative bacteria in the intensive care unit. Crit Care Med. 2010;38(8 Suppl):S345–51. doi: 10.1097/CCM.0b013e3181e6cbc5. [DOI] [PubMed] [Google Scholar]
- Martin YC, Jones PH, Perun TJ, Grudy WE, Bell S, Bower RR, Shipkowitz NL. Chemical modification of erythromycin antibiotics. 4. Structure-activity relations of erythromycin esters. J Med Chem. 1972;15:635–638. doi: 10.1021/jm00276a018. [DOI] [PubMed] [Google Scholar]
- Martinez JL, Sánchez MB, Martínez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev. 2009;33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- Mazzariol A, Cornaglia G, Nikaido H. Contributions of the AmpC β-lactamase and the AcrAB multidrug efflux system in intrinsic resistance of Escherichia coli K-12 to β-lactams. Antimicrob Agents Chemother. 2000;44:1387–1390. doi: 10.1128/aac.44.5.1387-1390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry LM, Cullinane JC, Levy SB. Transport of the lipophilic analog minocycline differs from that of tetracycline in susceptible and resistant Escherichia coli strains. Antimicrob Agents Chemother. 1982;22:791–799. doi: 10.1128/aac.22.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros AA, O’Brien TF, Rosenberg EY, Nikaido H. Loss of OmpC porin in a strain of Salmonella typhimurium causes increased resistance to cephalosporins during therapy. J Infect Dis. 1987;156:751–757. doi: 10.1093/infdis/156.5.751. [DOI] [PubMed] [Google Scholar]
- Misra R, Benson SA. Isolation and characterization of OmpC porin mutants with altered pore properties. J Bacteriol. 1988;170:528–533. doi: 10.1128/jb.170.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Linnell SK, Becnel Boyd L, Steffen D, Zechiedrich L. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009;53:235–241. doi: 10.1128/AAC.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C, Plésiat P, Jeannot K. A Two-Component Regulatory System Interconnects Resistance to Polymyxins, Aminoglycosides, Fluoroquinolones, and Beta-Lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2011;55:1211–1221. doi: 10.1128/AAC.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S. Multidrug efflux transporter, AcrB--the pumping mechanism. Curr Opin Struct Biol. 2008;18:459–465. doi: 10.1016/j.sbi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:5854–5858. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae R, Nakae T. Diffusion of aminoglycoside antibiotics across the outer membrane of Escherichia coli. Antimicrob Agents Chemother. 1982;22:554–559. doi: 10.1128/aac.22.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Ishida Y, Ohtsuka M, Kawato H, Yoshida K, Yokomizo Y, Hosono S, Ohta T, Hoshino K, Ishida H, Yoshida K, Renau TE, Léger R, Zhang JZ, Lee VJ, Watkins WJ. MexAB-OprM-specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 1: Discovery and early strategies for lead optimization. Bioorg Medic Chem Lett. 2003;13:4201–4204. doi: 10.1016/j.bmcl.2003.07.024. [DOI] [PubMed] [Google Scholar]
- Nestorovich EM, Danelon C, Winterhalter M, Bezrukov SM. Designed to penetrate: Time-resolved interaction of single antibiotic molecules with bacterial pores. Proc Natl Acad Sci U S A. 2002;99:9789–9794. doi: 10.1073/pnas.152206799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. The role of outer membrane and efflux pumps in the resistance of gram-negative bacteria. Can we improve drug access? Drug Resist Updat. 1998;1:93–98. doi: 10.1016/s1368-7646(98)80023-x. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Preventing drug access to targets: Cell surface permeability barriers and active efflux in bacteria. Semin Cell Dev Biol. 2001;12:215–223. doi: 10.1006/scdb.2000.0247. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Normark S. Sensitivity of Escherichia coli to various β-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic β-lactamases: A quantitative predictive treatment. Mol Microbiol. 1987;1:29–36. doi: 10.1111/j.1365-2958.1987.tb00523.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H, Rosenberg EY. Porin channels in Escherichia coli: Studies with liposomes reconstituted from purified proteins. J Bacteriol. 1983;153:241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Rosenberg EY, Foulds J. Porin channels in Escherichia coli: Studies with β-lactams in intact cells. J Bacteriol. 1983;153:232–240. doi: 10.1128/jb.153.1.232-240.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Liu W, Rosenberg EY. Outer membrane permeability and β-lactamase stability of dipolar ionic cephalosporins containing methoxyimino substituents. Antimicrob Agents Chemother. 1990;34:337–342. doi: 10.1128/aac.34.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Basina M, Nguyen V, Rosenberg EY. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side-chains. J Bacteriol. 1998;180:4686–4692. doi: 10.1128/jb.180.17.4686-4692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan E, Quinn T, Pagès JM, McCusker M, Piddock L, Fanning S. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar enteritidis: involvement of RamA and other global regulators. Antimicrob Agents Chemother. 2009;53:1080–1087. doi: 10.1128/AAC.01005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachón J, Vila J. Treatment of multiresistant Acinetobacter baumannii infections. Curr Opin Investig Drugs. 2009;10:150–156. [PubMed] [Google Scholar]
- Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2010;54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès JM, Amaral L. Mechanisms of drug efflux and strategies to combat them: challenging the efflux pump of Gram-negative bacteria. Biochim Biophys Acta. 2009;1794:826–833. doi: 10.1016/j.bbapap.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- Pagès JM, Lavigne JP, Leflon-Guibout V, Marcon E, Bert F, Noussair L, Nicolas-Chanoine MH. Efflux pump, the masked side of beta-lactam resistance in Klebsiella pneumoniae clinical isolates. PLoS One. 2009;4:e4817. doi: 10.1371/journal.pone.0004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès JM, Alibert-Franco S, Mahamoud A, Bolla JM, Davin-Regli A, Chevalier J, Garnotel E. Efflux Pumps of Gram-Negative Bacteria, a New Target for New Molecules. Curr Top Med Chem. 2010;8:1848–1857. doi: 10.2174/156802610793176620. [DOI] [PubMed] [Google Scholar]
- Peña C, Suarez C, Tubau F, Juan C, Moya B, Dominguez MA, Oliver A, Pujol M, Ariza J. Nosocomial outbreak of a non-cefepime-susceptible ceftazidime-susceptible Pseudomonas aeruginosa strain overexpressing MexXY-OprM and producing an integron-borne PSE-1 betta-lactamase. J Clin Microbiol. 2009;47:2381–2387. doi: 10.1128/JCM.00094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez A, Canle D, Latasa C, Poza M, Beceiro A, Tomás Mdel M, Fernández A, Mallo S, Pérez S, Molina F, Villanueva R, Lasa I, Bou G. Cloning, nucleotide sequencing, and analysis of the AcrAB-TolC efflux pump of Enterobacter cloacae and determination of its involvement in antibiotic resistance in a clinical isolate. Antimicrob Agents Chemother. 2007;51:3247–3253. doi: 10.1128/AAC.00072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Boto D, López-Portolés JA, Simón C, Valdezate S, Echeita MA. Study of the molecular mechanisms involved in high-level macrolide resistance of Spanish Campylobacter jejuni and Campylobacter coli strains. J Antimicrob Chemother. 2010;65:2083–2088. doi: 10.1093/jac/dkq268. [DOI] [PubMed] [Google Scholar]
- Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2007;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Plesiat P, Nikaido H. Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol Microbiol. 1992;6:1323–1333. doi: 10.1111/j.1365-2958.1992.tb00853.x. [DOI] [PubMed] [Google Scholar]
- Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005;49:479–487. doi: 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39:162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Pugsley AP, Schnaitman CA. Outer membrane proteins of Escherichia coli. VII. Evidence that bacteriophage-directed protein 2 functions as a pore. J Bacteriol. 1978;133:1181–1189. doi: 10.1128/jb.133.3.1181-1189.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn T, O’Mahony R, Baird AW, Drudy D, Whyte P, Fanning S. Multi-drug resistance in Salmonella enterica: efflux mechanisms and their relationships with the development of chromosomal resistance gene clusters. Curr Drug Targets. 2006;7:849–860. doi: 10.2174/138945006777709548. [DOI] [PubMed] [Google Scholar]
- Ricci V, Tzakas P, Buckley A, Piddock LJ. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob Agents Chemother. 2006;50:38–42. doi: 10.1128/AAC.50.1.38-42.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB. Unmet medical needs in antibacterial therapy. Biochem Pharmacol. 2006;71:991–995. doi: 10.1016/j.bcp.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Rice LB. The clinical consequences of antimicrobial resistance. Curr Opin Microbiol. 2009;12:476–481. doi: 10.1016/j.mib.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Roca I, Espinal P, Martí S, Vila J. First identification and characterization of an AdeABC-like efflux pump in Acinetobacter genomospecies 13TU. Antimicrob Agents Chemother. 2011;55:1285–1286. doi: 10.1128/AAC.01142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Immermann FW, Bradford PA. RT-PCR and statistical analyses of adeABC expression in clinical isolates of Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Microb Drug Resist. 2010;16:87–89. doi: 10.1089/mdr.2009.0131. [DOI] [PubMed] [Google Scholar]
- Schulz R, Vargiu AV, Collu F, Kleinekathöfer U, Ruggerone P. Functional rotation of the transporter AcrB: insights into drug extrusion from simulations. PLoS Comput Biol. 2010;6:e1000806. doi: 10.1371/journal.pcbi.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Miller MC, Brennan RG. Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. Embo J. 2004;23:2923–2930. doi: 10.1038/sj.emboj.7600288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A, Steinke P, Bohnert JA, Akova M, Jonas D, Kern WV. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J Antimicrob Chemother. 2006;57:344–348. doi: 10.1093/jac/dki446. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Trittler R, Bohnert JA, Kümmerer K, Pagès JM, Kern WV. Intracellular accumulation of linezolid in Escherichia coli, Citrobacter freundii and Enterobacter aerogenes: role of enhanced efflux pump activity and inactivation. J Antimicrob Chemother. 2007;59:1261–1264. doi: 10.1093/jac/dkl380. [DOI] [PubMed] [Google Scholar]
- Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by Darpin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennhauser G, Bukowska MA, Briand C, Grutter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol. 2009;389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Sigler A, Schubert P, Hillen W, Niederweis M. Permeation of tetracyclines through membranes of liposomes and Escherichia coli. Eur J Biochem. 2000;267:527–534. doi: 10.1046/j.1432-1327.2000.01026.x. [DOI] [PubMed] [Google Scholar]
- Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Govender N, Keddy KH Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa (GERMS-SA) Quinolone-resistant Salmonella Typhi in South Africa2003–2007. Epidemiol Infect. 2010;138:86–90. doi: 10.1017/S0950268809990331. [DOI] [PubMed] [Google Scholar]
- Sobel ML, Hocquet D, Cao L, Plésiat P, Poole K. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005;49:1782–1786. doi: 10.1128/AAC.49.5.1782-1786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnik-Isaac H, Weinberger M, Tabak M, Ben-David A, Shachar D, Yaron S. Quinolone resistance of Salmonella enterica serovar Virchow isolates from humans and poultry in Israel: evidence for clonal expansion. J Clin Microbiol. 2007;45:2575–2579. doi: 10.1128/JCM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CC, Yu EW. Ligand-transporter interaction in the AcrB multidrug efflux pump determined by fluorescence polarization assay. FEBS Lett. 2007;581:4972–4976. doi: 10.1016/j.febslet.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. Pseudomonas aeruginosa porin OprF exists in two different conformations. J Biol Chem. 2006;281:16220–16229. doi: 10.1074/jbc.M600680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara E, Nagano K, Nikaido H. Factors affecting the folding of Pseudomonas aeruginosa OprF porin into the one-domain open conformer. mBio. 2010;1:e00228–00210. doi: 10.1128/mBio.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. Expression of Multidrug Efflux Pump Genes acrAB-tolC, mdfA, and norE in Escherichia coli Clinical Isolates as a Function of Fluoroquinolone and Multidrug Resistance. Antimicrob Agents Chemother. 2011;55:921–914. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol. 2009;191:1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka Y, Chen C, Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci U S A. 2009;106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi DG, Suh GS, Nikaido H. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia coli. J Bacteriol. 1995;177:998–1007. doi: 10.1128/jb.177.4.998-1007.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Wang Q, Zgurskaya HI. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J Bacteriol. 2002;184:6499–6507. doi: 10.1128/JB.184.23.6499-6507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás M, Doumith M, Warner M, Turton JF, Beceiro A, Bou G, Livermore DM, Woodford N. Efflux pumps, OprD porin, AmpC beta-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother. 54:2219–2224. doi: 10.1128/AAC.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornroth-Horsefield S, Gourdon P, Horsefield R, Brive L, Yamamoto N, Mori H, Snijder A, Neutze R. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure. 2007;15:1663–1673. doi: 10.1016/j.str.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Tran QT, Dupont M, Lavigne JP, Chevalier J, Pagès JM, Sotto A, Davin-Regli A. Occurrence of efflux mechanism and cephalosporinase variant in a population of Enterobacter aerogenes and Klebsiella pneumoniae isolates producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2009;53:1652–1656. doi: 10.1128/AAC.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A, Kubo O, Miyamoto E, Yamana T. Physicochemical properties of β-lactam antibiotics: Oil-water distribution. J Pharm Sci. 1977;66:1675–1679. doi: 10.1002/jps.2600661205. [DOI] [PubMed] [Google Scholar]
- Tumah HN. Bacterial biocide resistance. J Chemother. 2009;21:5–15. [PubMed] [Google Scholar]
- Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1993;37:2255–2260. doi: 10.1128/aac.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bambeke F, Pagès JM, Lee VJ. Inhibitors of Bacterial Efflux Pumps as Adjuvants in Antibacterial Therapy and Diagnostic Tools for Detection of Resistance by Efflux. Frontiers in Anti-Infective Drug Discovery. 2010;1:138–175. doi: 10.2174/157489106777452692. [DOI] [PubMed] [Google Scholar]
- Vergidis PI, Falagas ME. Multidrug-resistant Gram-negative bacterial infections: the emerging threat and potential novel treatment options. Curr Opin Investig Drugs. 2008;9:176–183. [PubMed] [Google Scholar]
- Vettoretti L, Floret N, Hocquet D, Dehecq B, Plésiat P, Talon D, Bertrand X. Emergence of extensive-drug-resistant Pseudomonas aeruginosa in a French university hospital. Eur J Clin Microbiol Infect Dis. 2009;28:1217–1222. doi: 10.1007/s10096-009-0767-8. [DOI] [PubMed] [Google Scholar]
- Vettoretti L, Plésiat P, Muller C, El Garch F, Phan G, Attrée I, Ducruix A, Llanes C. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother. 2009;53:1987–1997. doi: 10.1128/AAC.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila J, Marti S, Sanchez-Cespedes J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumanii. J Antimicrob Chemother. 2007;59:1210–1215. doi: 10.1093/jac/dkl509. [DOI] [PubMed] [Google Scholar]
- Vu H, Nikaido H. Role of β-lactam hydrolysis in the mechanism of resistance of a β-lactamase-constitutive Enterobacter cloacae strain to expanded-spectrum β-lactams. Antimicrob Agents Chemother. 1985;27:393–398. doi: 10.1128/aac.27.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenlehner FM, Weidner W, Perletti G, Naber KG. Emerging drugs for bacterial urinary tract infections. Expert Opin Emerg Drugs. 2010;15:375–397. doi: 10.1517/14728214.2010.500613. [DOI] [PubMed] [Google Scholar]
- Yan M, Sahin O, Lin J, Zhang Q. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J Antimicrob Chemother. 2006;58:1154–1159. doi: 10.1093/jac/dkl412. [DOI] [PubMed] [Google Scholar]
- Yao XQ, Kenzaki H, Murakami S, Takada S. Drug export and allosteric coupling in a multidrug transporter revealed by molecular simulations. Nat Commun. 2010;1:117. doi: 10.1038/ncomms1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasufuku T, Shigemura K, Shirakawa T, Matsumoto M, Nakano Y, Tanaka K, Arakawa S, Kinoshita S, Kawabata M, Fujisawa M. Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli Strains clinically isolated from urinary tract infection patients. J Clin Microbiol. 2011;49:189–194. doi: 10.1128/JCM.00827-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F, Nikaido H. Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother. 1985;27:84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F, Zalman LS, Nikaido H. Purification and properties of Pseudomonas aeruginosa porin. J Biol Chem. 1983;258:2308–2314. [PubMed] [Google Scholar]
- Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: A crystallographic and site-directed mutagenesis study. J Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE., Jr Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- Zavascki AP, Carvalhaes CG, Picão RC, Gales AC. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev Anti Infect Ther. 2010;8:71–93. doi: 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- Ziha-Zarifi I, Llanes C, Köhler T, Péchère JC, Plésiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother. 1999;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]