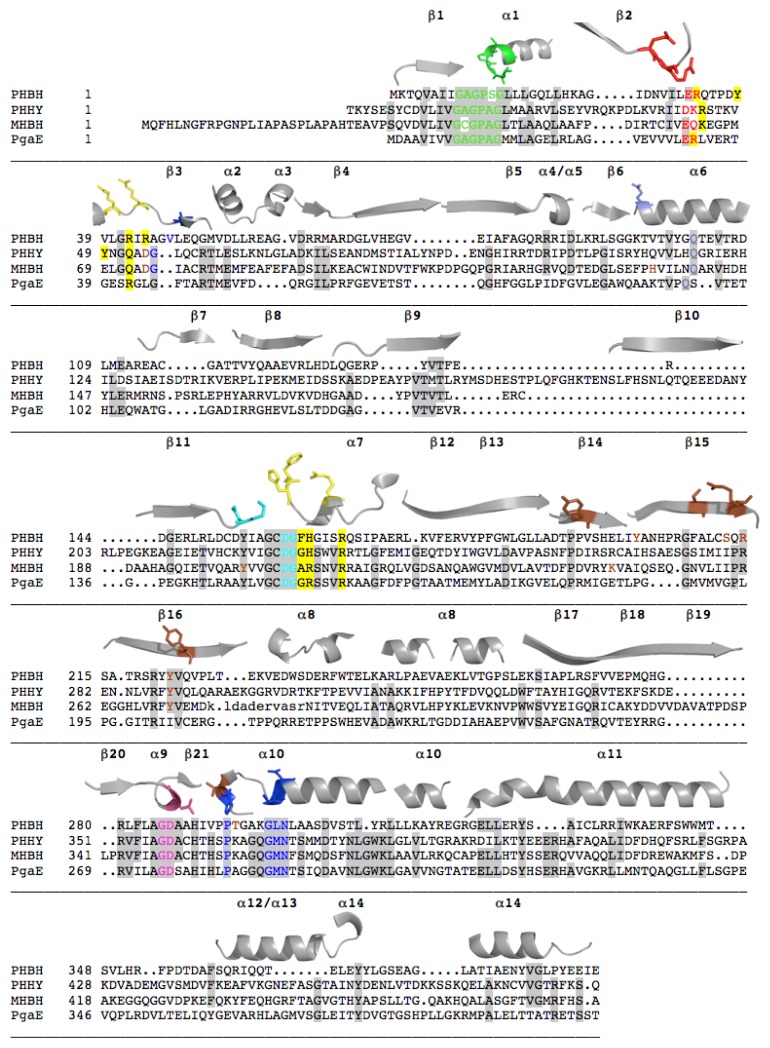
Figure 3.
Sequence and Structural Alignment Comparisons of Four Class A FPMOs. Amino acids shaded gray or otherwise highlighted are either fully or partially conserved. The colored residues represent important motifs found in or near the enzyme’s active site. The 3-dimensional location of the amino acids is ostensibly the same for each color of residue. The secondary structure elements comprising the structure and the position of each positionally conserved residue. These were rendered individually from the structure of PHBH (PDB ID, 1PBE). A three dimensional representation of these colored residues is shown in Figure 4. The brown residues indicate substrate binding residues (the yellow or periwinkle residues are involved in flavin movement and/or NADPH binding) and the remaining colored residues are primarily FAD binding motifs. Note that only domains I and II are aligned. The complete sequences for PHHY, PgaE and MHBH include domain III (Figure 2). Accession numbers for the sequences are, PHBH (AAA88455.1), PHHY (AAA34202.1), MHBH (BAF34928.1), PgaE (AAK57522.1). The structures used for the 3D structural alignment were from protein data bank files, 1PBE (PHBH), 1FOH (PHHY), 2DKH (MHBH), 2QA1 (PgaE).