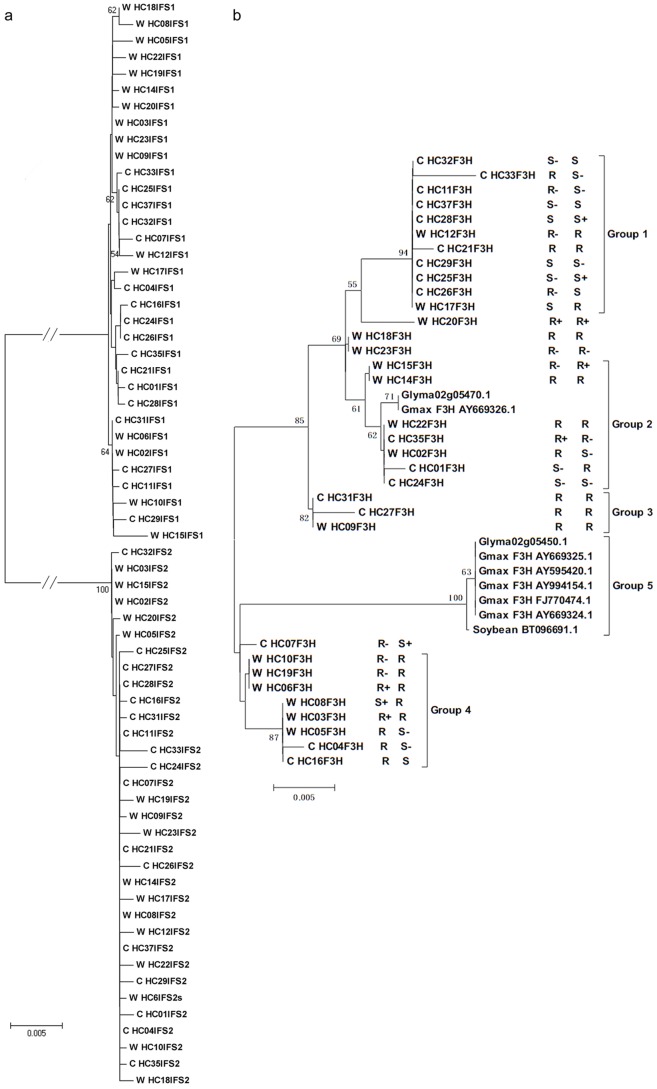
Figure 1. Phylogenetic trees of IFS1 and IFS2 (a), F3H2 (b).
The trees were estimated by neighbor-joining (NJ) method based on multiple CDS sequence alignments. CDS of F3H1 (Glyma02g05450.1) alleles were used as outgroup for soybean F3H2 in this study. The synthetic disease index (SDI) [32] was used in evaluation of soybean resistance to soybean mosaic virus. If the SDI was under 0, 20 and 35, the accession was classified as R+, R and R-. Meanwhile, we defined the accession as S+, S and S- respectively, if the SDI was above 70, 51 and 36.The resistance responses of 33 soybean accessions to SMV strains SC-3 and SC-7 were listed on the former and latter columns respectively. Bootstrap values >50% are indicated on the branches.