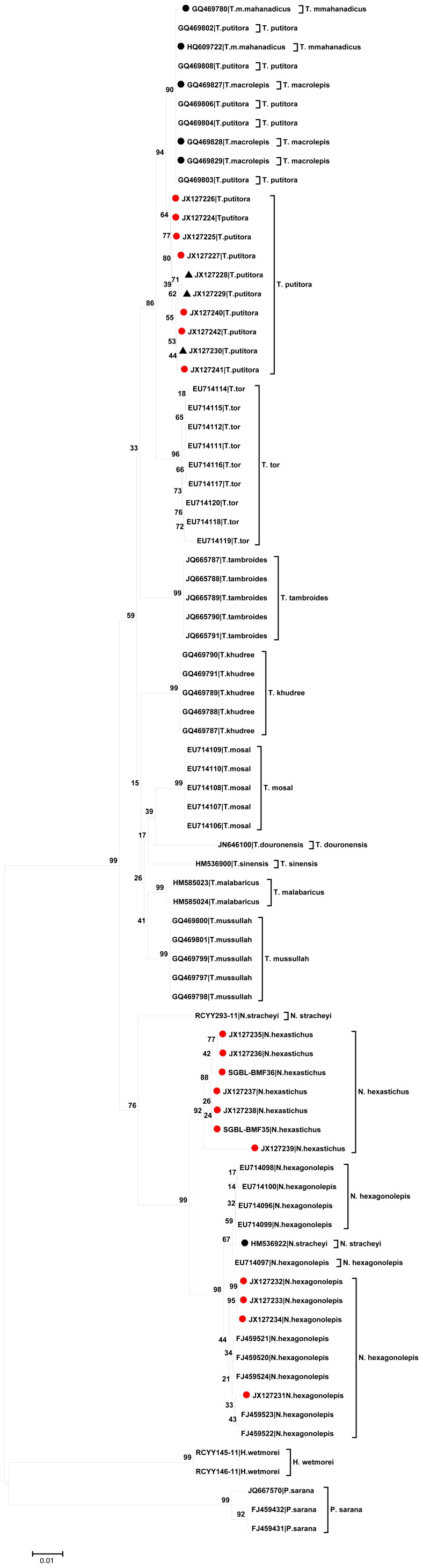
Figure 3. Neighbor Joining (NJ) tree developed using K2P distance among 76 COI sequences of mahseer.
In most cases the studied samples (marked as red dots) showed cohesive clustering with their conspecific database sequences. The samples morphologically identified as Tor progeneius (accession numbers JX127229, JX127230 and JX127228) clustered conspecific with developed sequences as well as database sequences of T. putitora. All samples of N. hexastichus clustered cohesive as the same species and distinct from N. hexagonolepis sequences. Few database sequences of 3 species revealed aberrant clustering {Neolissochilus stracheyi (accession number HM536922), Tor mosal mahanadicus (accession numbers HQ609722, GQ469780), Tor macrolepis (accession numbers GQ469827-29)}. • The numbers at the nodes are bootstrap values based on 1000 replications. • The specimens’ GenBank accession number and species name are shown for each taxon. • Red dots and black triangles correspond to the sequences developed in this study. Black triangles also correspond to the sequences of samples, although morphologically identified as Tor progeneius but were found conspecific with Tor putitora based on COI sequence data analysis. Black dots correspond to the cases of abnormal clustering.