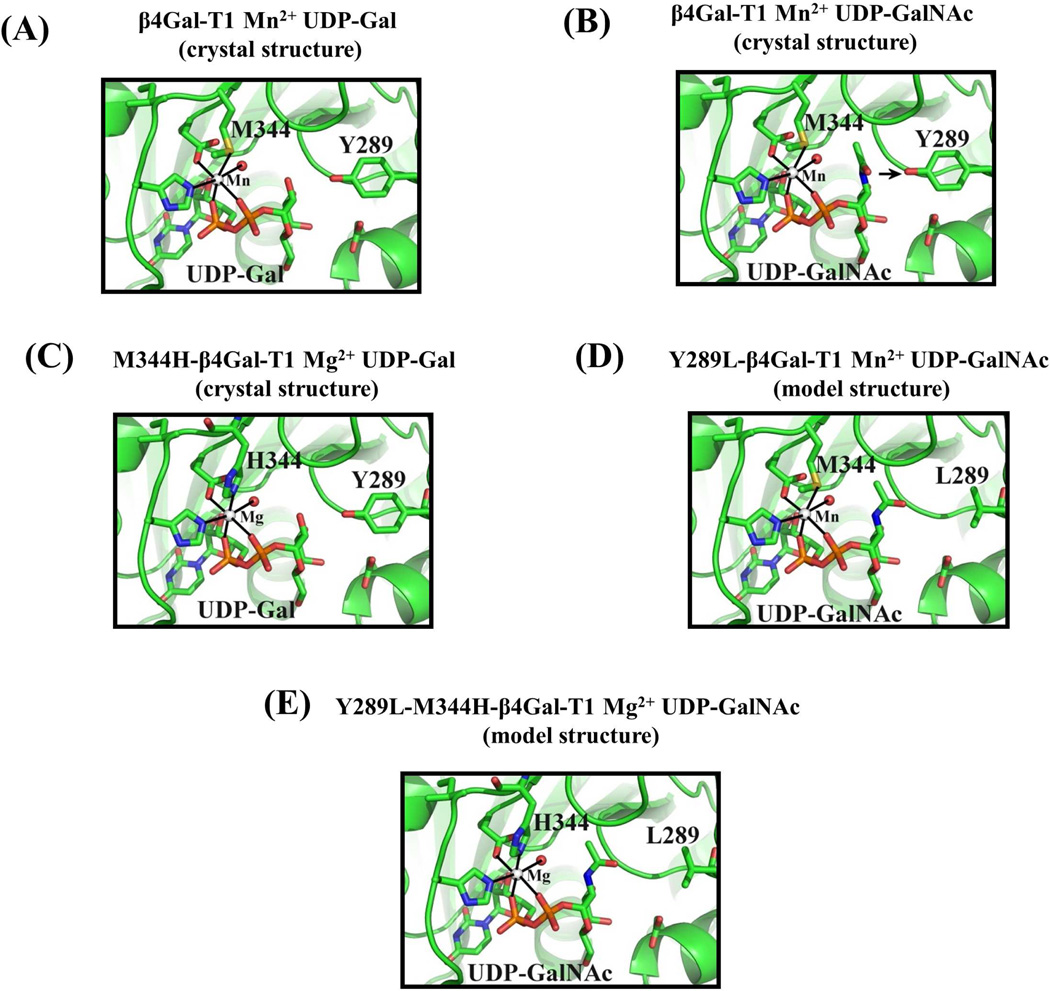
Figure 1.
A: In the crystal structure of the β4Gal-T1 complex with Mn2+ and UDP-Gal (pdb 1TVY), the side chain sulfur atom Sδof the Met-344 residue forms a coordination bond with the bound Mn2+ ion. B: In the crystal structure of the bovine β4Gal-T1 molecule that is in complex with UDP-GalNAc (pdb 1OQM), the N-acetyl moiety of the GalNAc sugar exhibits steric hindrance with the side chain of Tyr289. When the Tyr residue is mutated to Leu, resulting in the mutant Y289L-Gal-T1, it creates a cavity that can accommodate the N-acetyl moiety of GalNAc. The mutant enzyme Y289L-β4 GalT1 transfers GalNAc to GlcNAc (12). C: When the residue Met-344 is mutated to a His, a Mg2+ ion, in place of the Mn2+ ion, coordinates with the His-344 residue and activates the Met-344-His (M344H) mutant enzyme. D: Based on the crystal structure, the Y289L mutation has been modeled to show the space created between the L289 residue and the N-acetyl moiety of the GalNAc residue. E: The combination of both mutations, Y289L and M344H, has been modeled. The double mutant Y289L-M344H-β4 GalT1 is expected to transfer GalNAc sugars in the presence of Mg2+.