Figure 2.
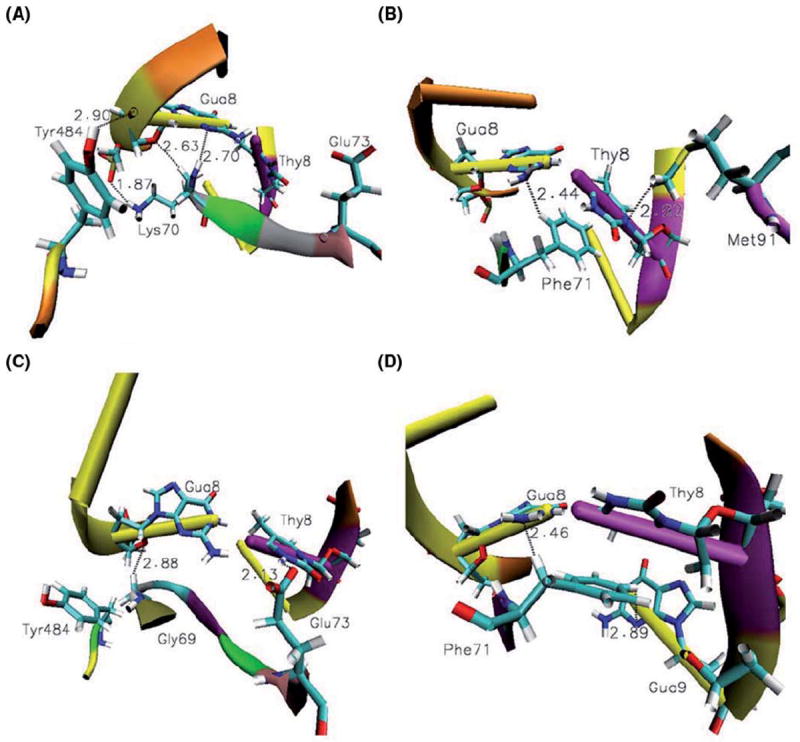
Recognition of the mismatched DNA by MutSα. (A) Tyr484 makes strong N-H…O hydrogen bonds with the mispaired guanine more than 83% of the simulation time. The side chain of Lys70 makes strong N-H…O and /or weak C-H…O, C-H…N hydrogen bonds with the mispaired guanine. Notice the orientation of the Glu73 away from the mismatched site which is prevalent more than 80% of the simulation time. (B) Phe71 and Met91 also probe the mismatched pair by strong and weak interactions present more than 80% of the simulation time. Recognition of cisplatin-DNA by MutSα. (C) The side chain of Glu73 makes strong N-H…O bonds with the mispaired thymine. Notice the orientation of Tyr484 away from the cross-linked DNA fragment, which is common almost half of the simulation time. Gly69 stabilizes the complex by weak C-H…O bonding with mispaired guanine. (D) Weak C-H…N bonds made by Phe71 with mismatched, platinum cross-linked guanine, Gua8, and the base adjacent to the mismatched thymine, Gua9.
