Figure 7. A mechanical model for homolog pairing.
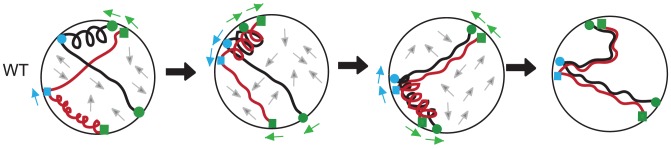
A hypothetical sequence of interactions between homologous chromosomes (shown as black or red) subjected to a coupled-spring oscillator (see text). In the sequence from left to right, homolog pairing becomes progressively stabilized as weak interactions are disrupted. The positive and negative forces of actin influence both homologs, with actin-based associations shown at telomeres (green; squares for the red chromosome and circles for the black homologous chromosome) and at interstitial sites (blue; squares for the red chromosome and circles for the black homologous chromosome). Arrows around the periphery of the nucleus indicate direction of movement for the telomeres (green) and the interstitial sites (blue). Grey arrows in the interior of the nucleus show “Brownian-like” motion/unknown forces on the chromosomes [105], [119]. (i) In wild-type cells, segments of chromosomes that are in closer proximity have axial segments that are more compact. (ii) Compact segments of homologous chromosomes interact. (iii) Movement of chromosome attachment points on the nuclear envelope results in stretching of segments that remove unstable interactions between chromosomes. (iv) Stable interactions between allelic loci are those achieved up to the point of dHJ resolution as established using the Cre/loxP assay [21].
